SML0778
UNC1999
≥98% (HPLC)
Sinônimo(s):
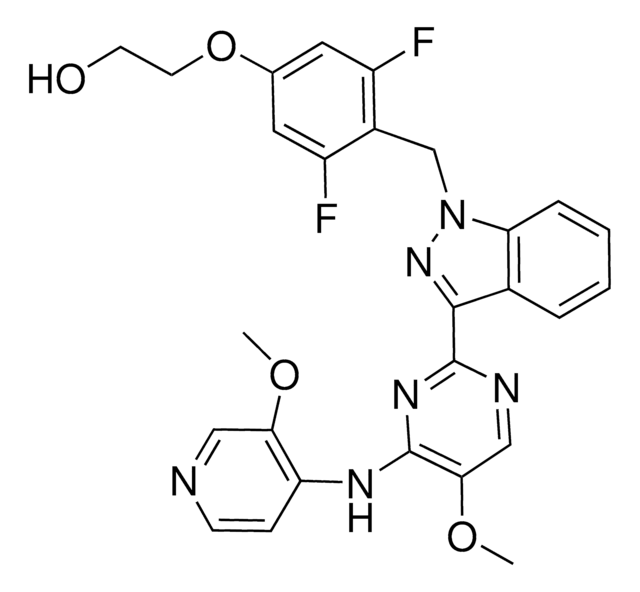
1-Isopropyl-6-(6-(4-isopropylpiperazin-1-yl)pyridin-3-yl)-N-((6-methyl-2-oxo-4-propyl-1,2-dihydropyridin-3-yl)methyl)-1H-indazole-4-carboxamide
About This Item
Produtos recomendados
Nível de qualidade
Ensaio
≥98% (HPLC)
Formulário
powder
cor
white to beige
solubilidade
DMSO: 15 mg/mL, clear
temperatura de armazenamento
2-8°C
cadeia de caracteres SMILES
CC(C)N1CCN(C2=NC=C(C3=CC4=C(C=NN4C(C)C)C(C(NCC5=C(CCC)C=C(C)NC5=O)=O)=C3)C=C2)CC1
InChI
1S/C33H43N7O2/c1-7-8-24-15-23(6)37-33(42)28(24)19-35-32(41)27-16-26(17-30-29(27)20-36-40(30)22(4)5)25-9-10-31(34-18-25)39-13-11-38(12-14-39)21(2)3/h9-10,15-18,20-22H,7-8,11-14,19H2,1-6H3,(H,35,41)(H,37,42)
chave InChI
DPJNKUOXBZSZAI-UHFFFAOYSA-N
Categorias relacionadas
Ações bioquímicas/fisiológicas
For full characterization details, please visit the UNC1999 probe summary on the Structural Genomics Consortium (SGC) website.
UNC2400 is the negative control for the active probe, UNC1999. To request a sample of the negative control from the SGC, click here.
To learn about other SGC chemical probes for epigenetic targets, visit sigma.com/sgc
Características e benefícios
Outras notas
produto relacionado
Código de classe de armazenamento
11 - Combustible Solids
Classe de risco de água (WGK)
WGK 3
Ponto de fulgor (°F)
Not applicable
Ponto de fulgor (°C)
Not applicable
Escolha uma das versões mais recentes:
Certificados de análise (COA)
Não está vendo a versão correta?
Se precisar de uma versão específica, você pode procurar um certificado específico pelo número do lote ou da remessa.
Já possui este produto?
Encontre a documentação dos produtos que você adquiriu recentemente na biblioteca de documentos.
Os clientes também visualizaram
Artigos
We offer a variety of small molecule research tools, such as transcription factor modulators, inhibitors of chromatin modifying enzymes, and agonists/antagonists for target identification and validation in gene regulation research; a selection of these research tools is shown below.
Nossa equipe de cientistas tem experiência em todas as áreas de pesquisa, incluindo Life Sciences, ciência de materiais, síntese química, cromatografia, química analítica e muitas outras.
Entre em contato com a assistência técnica