推薦產品
化驗
97%
mp
183-184 °C (lit.)
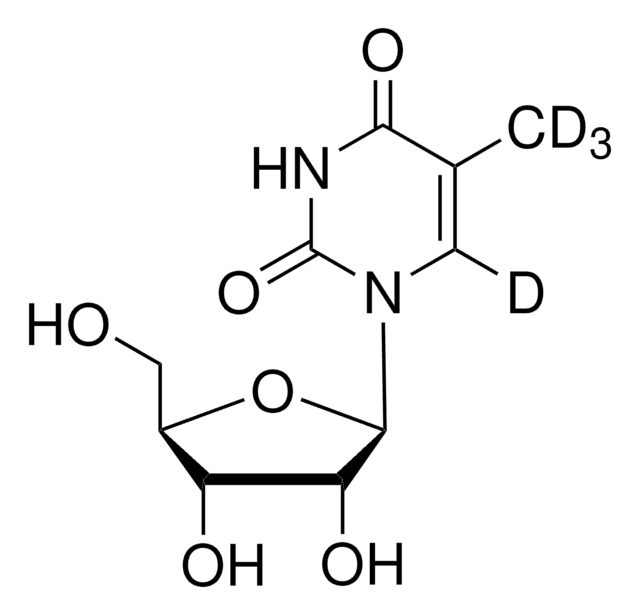
SMILES 字串
CC1=CN([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)C(=O)NC1=O
InChI
1S/C10H14N2O6/c1-4-2-12(10(17)11-8(4)16)9-7(15)6(14)5(3-13)18-9/h2,5-7,9,13-15H,3H2,1H3,(H,11,16,17)/t5-,6-,7-,9-/m1/s1
InChI 密鑰
DWRXFEITVBNRMK-JXOAFFINSA-N
尋找類似的產品? 前往 產品比較指南
儲存類別代碼
11 - Combustible Solids
水污染物質分類(WGK)
WGK 3
閃點(°F)
Not applicable
閃點(°C)
Not applicable
個人防護裝備
Eyeshields, Gloves, type N95 (US)
分析證明 (COA)
輸入產品批次/批號來搜索 分析證明 (COA)。在產品’s標籤上找到批次和批號,寫有 ‘Lot’或‘Batch’.。
客戶也查看了
H F Becker et al.
Journal of molecular biology, 274(4), 505-518 (1998-01-07)
Almost all transfer RNA molecules sequenced so far contain two universal modified nucleosides at positions 54 and 55, respectively: ribothymidine (T54) and pseudouridine (psi 55). To identify the tRNA elements recognized by tRNA:m5uridine-54 methyltransferase and tRNA:pseudouridine-55 synthase from the yeast
Benoit Desmolaize et al.
Nucleic acids research, 39(21), 9368-9375 (2011-08-10)
Methyltransferases that use S-adenosylmethionine (AdoMet) as a cofactor to catalyse 5-methyl uridine (m(5)U) formation in tRNAs and rRNAs are widespread in Bacteria and Eukaryota, and are also found in certain Archaea. These enzymes belong to the COG2265 cluster, and the
Gregory E R Gordon et al.
Journal of biotechnology, 151(1), 108-113 (2010-11-30)
This paper describes a high yielding coupled enzymatic reaction using Bacillus halodurans purine nucleoside phosphorylase (PNP) and E. coli uridine phosphorylase (UP) for synthesis of 5-methyluridine (5-MU) by transglycosylation. Key parameters such as reaction temperature, pH, reactant loading, reactor configuration
Marcus J O Johansson et al.
RNA (New York, N.Y.), 8(3), 324-335 (2002-05-11)
A 5-methyluridine (m(5)U) residue at position 54 is a conserved feature of bacterial and eukaryotic tRNAs. The methylation of U54 is catalyzed by the tRNA(m5U54)methyltransferase, which in Saccharomyces cerevisiae is encoded by the nonessential TRM2 gene. In this study, we
Clive Persaud et al.
Biochemical and biophysical research communications, 392(2), 223-227 (2010-01-14)
Ribosomal RNAs (rRNAs) from all kingdoms contain a variety of post-transcriptional modifications and these are typically clustered in the functional centers of the ribosome. The functions of two bases in the 23S rRNA of Escherichia coli that are post-transcriptionally modified
我們的科學家團隊在所有研究領域都有豐富的經驗,包括生命科學、材料科學、化學合成、色譜、分析等.
聯絡技術服務