919896
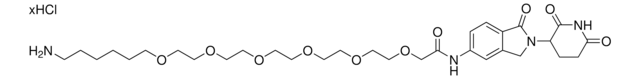
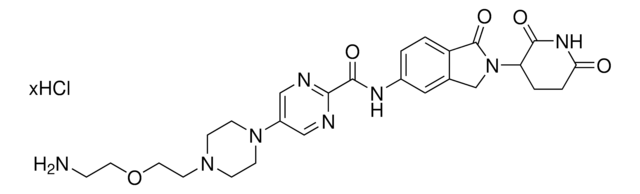
C5 Lenalidomide-dipiperazine-NH2 hydrochloride
Sinónimos:
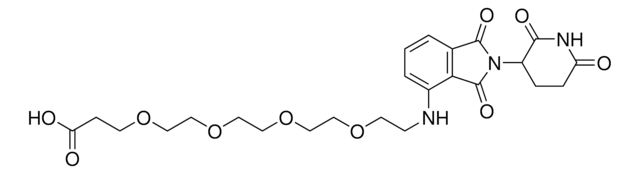
4-(4-(2-Aminoethyl)piperazine-1-carbonyl)-N-(2-(2,6-dioxopiperidin-3-yl)-1-oxoisoindolin-5-yl)piperazine-1-carboxamide, C5 Lenalidomide conjugate, Crosslinker−E3 Ligase ligand conjugate, Protein degrader building block for PROTAC® research, Template for synthesis of targeted protein degrader
About This Item
Productos recomendados
ligand
C5 Lenalidomide
Nivel de calidad
formulario
solid
idoneidad de la reacción
reactivity: carboxyl reactive
reagent type: ligand-linker conjugate
temp. de almacenamiento
2-8°C
cadena SMILES
O=C(N(CC1)CCN1C(NC2=CC(C3)=C(C=C2)C(N3C(C4=O)CCC(N4)=O)=O)=O)N5CCN(CC5)CCN.Cl
Aplicación
Otras notas
Portal: Building PROTAC® Degraders for Targeted Protein Degradation
Targeted Protein Degradation by Small Molecules
Small-Molecule PROTACS: New Approaches to Protein Degradation
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Información legal
Producto relacionado
Código de clase de almacenamiento
11 - Combustible Solids
Clase de riesgo para el agua (WGK)
WGK 3
Punto de inflamabilidad (°F)
Not applicable
Punto de inflamabilidad (°C)
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Artículos
Protein Degrader Building Blocks are a collection of crosslinker-E3 ligand conjugates with a pendant functional group for covalent linkage to a target ligand.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico
![Exo-Phenyl Kwon [2.2.1] Bicyclic Phosphine 95% (HPLC)](/deepweb/assets/sigmaaldrich/product/structures/477/026/5255f657-4af5-47da-9839-86b94d92129f/640/5255f657-4af5-47da-9839-86b94d92129f.png)
![Exo-4-anisole Kwon [2.2.1] bicyclic phosphine](/deepweb/assets/sigmaaldrich/product/structures/114/753/2a544671-b0e0-4556-8dc3-46c126d6c8ab/640/2a544671-b0e0-4556-8dc3-46c126d6c8ab.png)