C24604
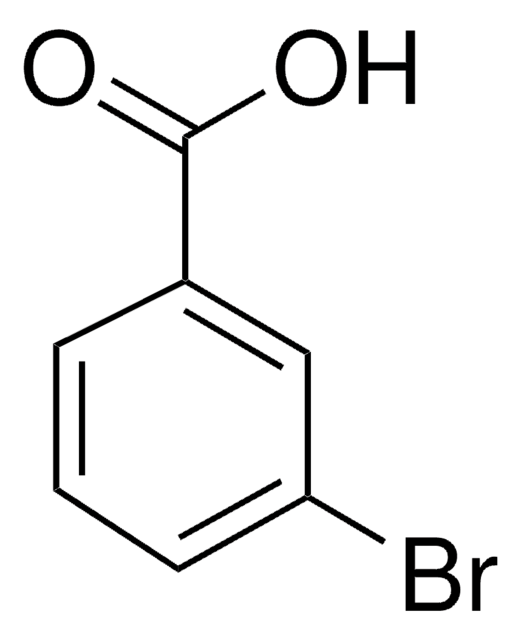
3-Chlorobenzoic acid
ReagentPlus®, ≥99%
Sinónimos:
m-Chlorobenzoic acid
About This Item
Productos recomendados
product line
ReagentPlus®
assay
≥99%
form
powder
mp
153-157 °C (lit.)
SMILES string
OC(=O)c1cccc(Cl)c1
InChI
1S/C7H5ClO2/c8-6-3-1-2-5(4-6)7(9)10/h1-4H,(H,9,10)
InChI key
LULAYUGMBFYYEX-UHFFFAOYSA-N
¿Está buscando productos similares? Visita Guía de comparación de productos
Categorías relacionadas
Legal Information
signalword
Warning
hcodes
Hazard Classifications
Eye Irrit. 2 - Skin Irrit. 2
Storage Class
11 - Combustible Solids
wgk_germany
WGK 3
ppe
dust mask type N95 (US), Eyeshields, Gloves
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico