MBD0042
Escherichia coli FISH probe - Cy3
Probe for fluorescence in situ hybridization (FISH)
Anmeldenzur Ansicht organisationsspezifischer und vertraglich vereinbarter Preise
Alle Fotos(3)
About This Item
UNSPSC-Code:
12352200
NACRES:
NA.55
Empfohlene Produkte
Qualitätsniveau
Methode(n)
FISH: suitable
Fluoreszenz
λex 550 nm; λem 570 nm
Versandbedingung
dry ice
Lagertemp.
−20°C
Allgemeine Beschreibung
Fluorescent In Situ Hybridization technique (FISH) is based on the hybridization of fluorescent labeled oligonucleotide probe to a specific complementary DNA or RNA sequence in whole and intact cells.1 Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples.2
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems 6) and plants7. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest.
Escherichia coli is a gram negative, facultative aerobic, rod-shaped coliform bacterium. E. coli colonizes the infant gut within hours of birth and establishes itself as the most abundant facultative anaerobe of the human intestinal microflora for the remainder of life, equipped with the abilities to grow in the ever-changing environment in the gut and cope with the mammalian host interaction.8,9 Nevertheless, E. coli can survive in many different ecological habitats, including abiotic environments, and is considered a highly versatile species. Known habitats of E. coli include soil, water, sediment, and food. Some strains of E. coli have evolved and adapted to a pathogenic lifestyle and can cause different disease pathologies.10
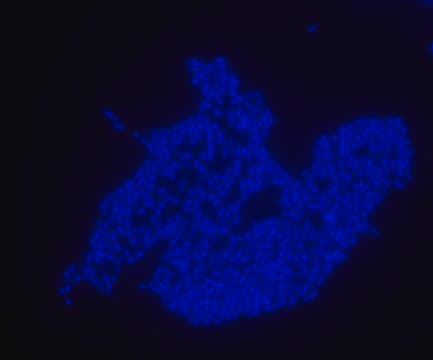
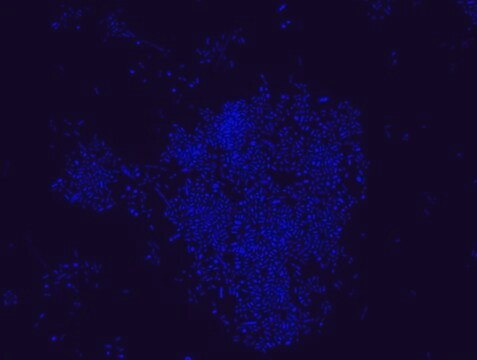
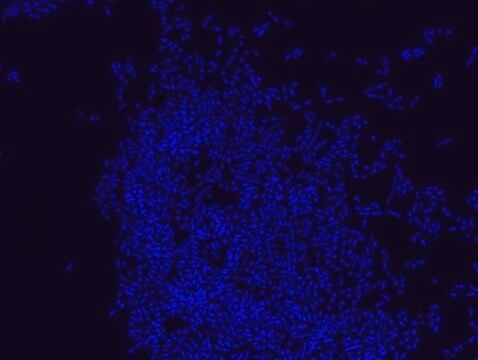
Escherichia coli probe specifically recognizes Escherichia coli cells.
Yet there are some reports that describe recognition of other bacteria with this probe, such as, Shigella boydii, Citrobacter davisae, Citrobacter lapagei, Citrobacter neteri 11 and Klebsiella pneumoniae 12.
FISH technique was successfully used to identify E.coli with the probe in various samples such as pure culture (as described in the figure legends and11,13), large and small intestines samples14-16, fecal samples17-21, colonic biopsies18, urine samples, bladder and kidney sections embedded in paraffin22 and in E.coli biofilm23.
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems 6) and plants7. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest.
Escherichia coli is a gram negative, facultative aerobic, rod-shaped coliform bacterium. E. coli colonizes the infant gut within hours of birth and establishes itself as the most abundant facultative anaerobe of the human intestinal microflora for the remainder of life, equipped with the abilities to grow in the ever-changing environment in the gut and cope with the mammalian host interaction.8,9 Nevertheless, E. coli can survive in many different ecological habitats, including abiotic environments, and is considered a highly versatile species. Known habitats of E. coli include soil, water, sediment, and food. Some strains of E. coli have evolved and adapted to a pathogenic lifestyle and can cause different disease pathologies.10
Escherichia coli probe specifically recognizes Escherichia coli cells.
Yet there are some reports that describe recognition of other bacteria with this probe, such as, Shigella boydii, Citrobacter davisae, Citrobacter lapagei, Citrobacter neteri 11 and Klebsiella pneumoniae 12.
FISH technique was successfully used to identify E.coli with the probe in various samples such as pure culture (as described in the figure legends and11,13), large and small intestines samples14-16, fecal samples17-21, colonic biopsies18, urine samples, bladder and kidney sections embedded in paraffin22 and in E.coli biofilm23.
Anwendung
Probe for fluorescence in situ hybridization (FISH),recognizes Escherichia coli cells
Leistungsmerkmale und Vorteile
- Visualize, identify and isolate Escherichia coli cells.
- Observe native E. coli cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of E. coli in bacterial mixed population.
- Specific, sensitive and robust identification even when E. coli is in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on E.coli morphology and allows to study biofilm architecture.
- Identify E.coli in clinical samples such as, urine samples, bladder and kidney sections (formalin-fixed paraffin-embedded (FFPE) samples), fecal samples and colon tissue.
- The ability to detect E.coli in its natural habitat is an essential tool for studying host-microbiome interaction.
Lagerklassenschlüssel
12 - Non Combustible Liquids
WGK
nwg
Flammpunkt (°F)
Not applicable
Flammpunkt (°C)
Not applicable
Analysenzertifikate (COA)
Suchen Sie nach Analysenzertifikate (COA), indem Sie die Lot-/Chargennummer des Produkts eingeben. Lot- und Chargennummern sind auf dem Produktetikett hinter den Wörtern ‘Lot’ oder ‘Batch’ (Lot oder Charge) zu finden.
Besitzen Sie dieses Produkt bereits?
In der Dokumentenbibliothek finden Sie die Dokumentation zu den Produkten, die Sie kürzlich erworben haben.
Arthur C Ouwehand et al.
Microbiology and immunology, 48(7), 497-500 (2004-07-24)
The fecal and mucosal microbiota of infants with rectal bleeding and the fecal microbiota of healthy age-matched controls were investigated by fluorescent in situ hybridization. Bifidobacteria were the main genus in both the feces and mucosa. The other genera tested
L K Poulsen et al.
Infection and immunity, 62(11), 5191-5194 (1994-11-01)
Fluorescent oligonucleotide probes targeting rRNA were used to develop an in situ hybridization technique by which the spatial distribution of Escherichia coli in the large intestines of streptomycin-treated mice was determined. Single E. coli cells were identified in thin frozen
Sahar Soleimani et al.
Applied microbiology and biotechnology, 97(3), 1093-1102 (2012-09-11)
Biofilms of selected bacteria strains were previously used on metal coupons as a protective layer against microbiologically influenced corrosion of metals. Unlike metal surfaces, concrete surfaces present a hostile environment for growing a protective biofilm. The main objective of this
D Grimm et al.
Applied and environmental microbiology, 64(7), 2686-2690 (1998-07-02)
Based on comparative sequence analysis, we have designed an oligonucleotide probe complementary to a region of 16S rRNA of Legionella pneumophila which allows the differentiation of L. pneumophila from other Legionella species without cultivation. The specificity of the new probe
Stuart C Smith et al.
European journal of nutrition, 45(6), 335-341 (2006-06-10)
Changes in the composition of gastrointestinal microbiota by dietary interventions using pro- and prebiotics provide opportunity for improving health and preventing disease. However, the capacity of lupin kernel fiber (LKFibre), a novel legume-derived food ingredient, to act as a prebiotic
Unser Team von Wissenschaftlern verfügt über Erfahrung in allen Forschungsbereichen einschließlich Life Science, Materialwissenschaften, chemischer Synthese, Chromatographie, Analytik und vielen mehr..
Setzen Sie sich mit dem technischen Dienst in Verbindung.