909629
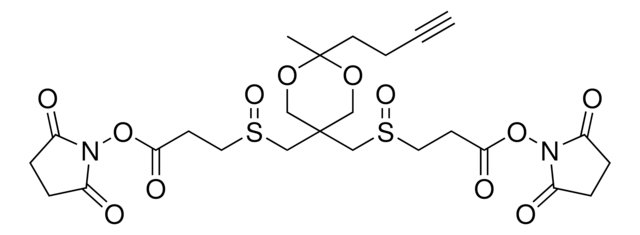
Azide-A-DSBSO crosslinker
≥95%
Synonyme(s) :
Azide-tagged acid-cleavable disuccinimidyl bissulfoxide, Bis(2,5-dioxopyrrolidin-1-yl) 3,3′-((2-(3-azidopropyl)-2-methyl-1,3-dioxane-5,5-diyl)bis(methylenesulfinyl))dipropionate, Mass spectrometry-cleavable crosslinker for studying protein-protein interations
About This Item
Produits recommandés
Niveau de qualité
Pureté
≥95%
Forme
powder
Disponibilité
available only in USA
Température de stockage
2-8°C
Application
Azide-A-DSBSO possesses two N-hydroxysuccinimide (NHS) ester groups for targeting amines, a ∼14 Å spacer length, two symmetrical acid-cleavable C-S bonds, and a central bioorthogonal azide tag. After reacting with lysine (Lys) residues, the azide tag permits selective enrichment of crosslinked proteins or peptides with a biotin-conjugated alkyne probe to enhance their detection. Additionally, the post-cleavage spacer yields tagged peptides for unambiguous identification by collision-induced dissociation in tandem MS.
Azide-A-DSBSO Crosslinker has mapped in vivo PPIs for target protein complexes and whole proteomes from living cells. It will also be a useful proteomics tool in the quest for targeting ″undruggable″ protein targets.
Autres remarques
A New in Vivo Cross-linking Mass Spectrometry Platform to Define Protein-Protein Interactions in Living Cells
Synthesis of two new enrichable and MS-cleavable cross-linkers to define protein–protein interactions by mass spectrometry
Informations légales
Produit(s) apparenté(s)
Code de la classe de stockage
4.1B - Flammable solid hazardous materials
Classe de danger pour l'eau (WGK)
WGK 3
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Articles
Sulfoxide-containing MS-cleavable cross-linkers capture protein-protein interactions in native cellular environments, aiding PPI identification.
Sulfoxide-containing MS-cleavable cross-linkers capture protein-protein interactions in native cellular environments, aiding PPI identification.
Sulfoxide-containing MS-cleavable cross-linkers capture protein-protein interactions in native cellular environments, aiding PPI identification.
Sulfoxide-containing MS-cleavable cross-linkers capture protein-protein interactions in native cellular environments, aiding PPI identification.
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique