MBD0059
Alphaproteobacteria FISH probe-Cy3
Probe for fluorescence in situ hybridization (FISH), 20 μM in water
Synonym(s):
ALF968
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
UNSPSC Code:
12352200
NACRES:
NA.55
Recommended Products
Quality Level
concentration
20 μM in water
fluorescence
λex 550 nm; λem 570 nm (Cy3)
shipped in
dry ice
storage temp.
-10 to -25°C
General description
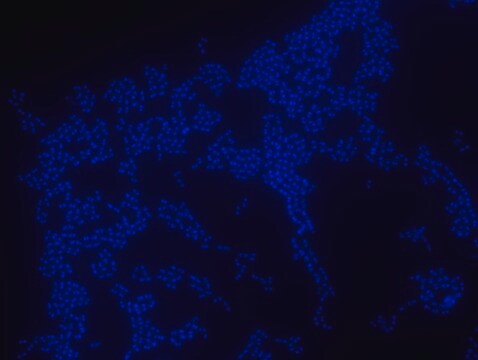
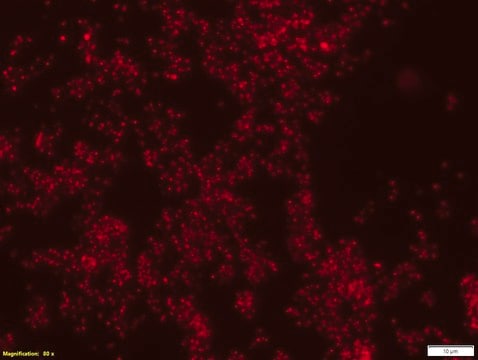
FISH (Fluorescent in Situ Hybridization) technique is based on the hybridization of fluorescently labeled oligonucleotide probes to a specific complementary DNA or RNA sequence in whole and intact cells. Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples. FISH can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood and other tissues), microbial ecology (solid biofilms and aquatic systems) and plants. Alphaproteobacteria FISH probe – Cy3 (MBD0059) specifically recognizes Alphaproteobacteria cells. Alphaproteobacteria is the second largest class of bacteria within the gram-negative phylum Proteobacteria, with a high level of functional diversity among its eight major orders: Caulobacterales, Rhizobiales, Rhodobacterales, Pelagibacterales, Sphingomonadales, Rhodospirillales, Holosporales and Rickettsiales. Mitochondria are thought to have evolved from Alphaproteobacteria. Between 20-50% of bacterioplankton cells in the ocean belong to the Alphaproteobacteria class, and Alphaproteobacteria is also abundant in soil and freshwater environments. Rhizobiales is a soil-based plant symbiont Alphaproteobacteria widely studied in agricultural research and Rhodobacterales is an example of a purple photosynthetic Alphaproteobacteria bacteria. Certain genera of Alphaproteobacteria, such as Brucella, are common human pathogens. One species of Alphaproteobacteria, Paracoccus yeei, was correlated to opportunistic infections in humans, including peritonitis,myocarditis, and dermatologic lesions. Interestingly, several types of Alphaproteobacteria have been shown to divide asymmetrically, a trait that may contribute to this class’s heterogeneity and adaptability to a wide variety of microbiome environments. The FISH probe has been used to recognize many Alphaproteobacteria species, such as Magnetospirillum gryphiswaldense, a spiral-shaped magnetotactic bacteria first isolated from river sediment in 1990. In addition, the FISH probe has been used to detect Alphaproteobacteria from a wide variety of environmental microbial samples, including bacterial populations isolated from snow, wastewater treatment plants, lichen and sphagnum moss samples, seawater, and freshwater sediments.
Application
Probe for fluorescence in situ hybridization (FISH), recognizes Alphaproteobacteria cells (except for Rickettsiales)
Features and Benefits
- Visualize, identify and isolate Alphaproteobacteria cells.
- Observe native Alphaproteobacteria cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of Alphaproteobacteria in bacterial mixed populations.
- Specific, sensitive and robust identification even when Alphaproteobacteria is in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on Alphaproteobacteria morphology
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria
Cardinale, et al.
FEMS Microbiology Ecology, 66(1), 63-71 (2008)
Genome streamlining in a cosmopolitan oceanic bacterium
Science, 309(5738), 1242-1245 (2005)
An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins
eLife, 8 (2019)
First comprehensively documented case of Paracoccus yeei infection in a human
Journal of Clinical Microbiology, 42(7), 3366-3368 (2004)
A DNA microarray platform based on direct detection of rRNA for characterization of freshwater sediment-related prokaryotic communities
Peplies, et al.
Applied and Environmental Microbiology, 72(7), 4829-4838 (2006)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service