MBD0041
Akkermansia muciniphila FISH probe - Cy3
Probe for fluorescence in situ hybridization (FISH),20 μM in water
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
UNSPSC Code:
12352200
NACRES:
NA.55
Recommended Products
Quality Level
technique(s)
FISH: suitable
fluorescence
λex 550 nm; λem 570 nm (Cy3)
shipped in
dry ice
storage temp.
−20°C
General description
Fluorescent In Situ Hybridization technique (FISH) is based on the hybridization of fluorescent labeled oligonucleotide probe to a specific complementary DNA or RNA sequence in whole and intact cells.1 Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples.2
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems6) and plants7. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest.
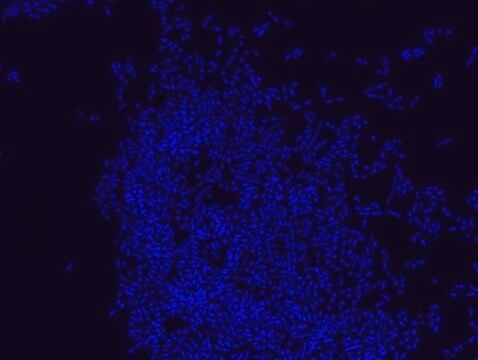
Akkermansia muciniphila probe specifically recognizes Akkermansia muciniphila cells.
Akkermansia muciniphila is a gram negative, oval shaped, non-motile, non-spore forming strictly anaerobic bacteria.8 A.muciniphila inhabits the gastrointestinal tracts of more than 90% of adults and constitutes 1 to 4% of the fecal microbiota.9 It is one of the top 20 most abundant species detectable in the human gut.10
The mucus layer of the human intestine is a niche which is colonized by specific bacteria such as A. muciniphila. A. muciniphila is able to degrade mucin, a key mucus component, using the enzymes sialidase and fucosidase, and utilize it as a source of carbon and nitrogen.11 Consequently, the host produces additional mucus while the bacterium produces oligosaccharides and Short Chain Fatty Acids (SCFAs) that can be utilized by the host and trigger the immune system. An additional protective effect of the SCFA is stimulation of mucus-associated microbiota growth, that serves as a barrier against penetration of pathogens to intestinal cells.9,12
It was found that A.muciniphila abundance in the gut was correlated to a healthy intestine and inversely correlated to many disease conditions.11 In comparison to healthy controls, A.muciniphila levels were low in patients with intestinal disorders, such as inflammatory bowel disease (IBD), but also in other conditions, such as autism, atopy, and obesity.11,13-16 Therefore, the level of A.muciniphila was suggested to serve as a biomarker for healthy intestine.17
A. muciniphila is a promising potential probiotic that can be administrated for the treatment of diseases such as, colitis, metabolic syndromes, immune diseases and cancer.10
FISH technique was successfully used to identify A.muciniphila with the probe in various samples such as pure culture (as described in the figure legends and18), fecal samples 19-21, gut lumen content 22, appendix samples 23, cecum content and tissue24,25 and colon tissue26. The probe can also be used for FISH coupled with flow cytometry (FCM-FISH)19,20,21 and FISH combined with Raman microspectroscopy 24.
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems6) and plants7. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest.
Akkermansia muciniphila probe specifically recognizes Akkermansia muciniphila cells.
Akkermansia muciniphila is a gram negative, oval shaped, non-motile, non-spore forming strictly anaerobic bacteria.8 A.muciniphila inhabits the gastrointestinal tracts of more than 90% of adults and constitutes 1 to 4% of the fecal microbiota.9 It is one of the top 20 most abundant species detectable in the human gut.10
The mucus layer of the human intestine is a niche which is colonized by specific bacteria such as A. muciniphila. A. muciniphila is able to degrade mucin, a key mucus component, using the enzymes sialidase and fucosidase, and utilize it as a source of carbon and nitrogen.11 Consequently, the host produces additional mucus while the bacterium produces oligosaccharides and Short Chain Fatty Acids (SCFAs) that can be utilized by the host and trigger the immune system. An additional protective effect of the SCFA is stimulation of mucus-associated microbiota growth, that serves as a barrier against penetration of pathogens to intestinal cells.9,12
It was found that A.muciniphila abundance in the gut was correlated to a healthy intestine and inversely correlated to many disease conditions.11 In comparison to healthy controls, A.muciniphila levels were low in patients with intestinal disorders, such as inflammatory bowel disease (IBD), but also in other conditions, such as autism, atopy, and obesity.11,13-16 Therefore, the level of A.muciniphila was suggested to serve as a biomarker for healthy intestine.17
A. muciniphila is a promising potential probiotic that can be administrated for the treatment of diseases such as, colitis, metabolic syndromes, immune diseases and cancer.10
FISH technique was successfully used to identify A.muciniphila with the probe in various samples such as pure culture (as described in the figure legends and18), fecal samples 19-21, gut lumen content 22, appendix samples 23, cecum content and tissue24,25 and colon tissue26. The probe can also be used for FISH coupled with flow cytometry (FCM-FISH)19,20,21 and FISH combined with Raman microspectroscopy 24.
Application
Probe for fluorescence in situ hybridization (FISH),recognizes Akkermansia muciniphila cells
Features and Benefits
- Visualize, identify and isolate Akkermansia muciniphila cells.
- Observe native A. muciniphila cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of A.muciniphila in bacterial mixed population.
- Specific, sensitive and robust identification even when A. muciniphila is in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on A.muciniphila morphology.
- Identify A.muciniphila in clinical samples such as, gut lumen content, appendix samples (formalin-fixed paraffin-embedded (FFPE) samples), fecal samples and colon tissue.
- The ability to detect A.muciniphila in its natural habitat is an essential tool for studying host-microbiome interaction.
Storage Class Code
12 - Non Combustible Liquids
WGK
nwg
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Lv Wang et al.
Applied and environmental microbiology, 77(18), 6718-6721 (2011-07-26)
Gastrointestinal disturbance is frequently reported for individuals with autism. We used quantitative real-time PCR analysis to quantify fecal bacteria that could influence gastrointestinal health in children with and without autism. Lower relative abundances of Bifidobacteria species and the mucolytic bacterium
Noora Ottman et al.
Best practice & research. Clinical gastroenterology, 31(6), 637-642 (2018-03-24)
The discovery of Akkermansia muciniphila has opened new avenues for the use of this abundant intestinal symbiont in next generation therapeutic products, as well as targeting microbiota dynamics. A. muciniphila is known to colonize the mucosal layer of the human intestine
David Berry et al.
The ISME journal, 6(11), 2091-2106 (2012-05-11)
Human inflammatory bowel disease and experimental colitis models in mice are associated with shifts in intestinal microbiota composition, but it is unclear at what taxonomic/phylogenetic level such microbiota dynamics can be indicative for health or disease. Here, we report that
Husen Zhang et al.
Proceedings of the National Academy of Sciences of the United States of America, 106(7), 2365-2370 (2009-01-24)
Recent evidence suggests that the microbial community in the human intestine may play an important role in the pathogenesis of obesity. We examined 184,094 sequences of microbial 16S rRNA genes from PCR amplicons by using the 454 pyrosequencing technology to
David Berry et al.
Proceedings of the National Academy of Sciences of the United States of America, 112(2), E194-E203 (2015-01-01)
Microbial communities are essential to the function of virtually all ecosystems and eukaryotes, including humans. However, it is still a major challenge to identify microbial cells active under natural conditions in complex systems. In this study, we developed a new
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service