MBD0033
Eubacteria FISH probe - Cy3
Probe for fluorescence in situ hybridization (FISH)
Synonym(s):
EUB338
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
UNSPSC Code:
12352200
NACRES:
NA.31
Recommended Products
Quality Level
form
liquid
concentration
20 μM
technique(s)
FISH: suitable
shipped in
dry ice
storage temp.
−20°C
General description
Fluorescent In Situ Hybridization technique (FISH) is based on the hybridization of fluorescent labeled oligonucleotide probe to a specific complementary DNA or RNA sequence in whole and intact cells.1 Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples.2
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems6) and plants7.
Prokaryotic single cell life forms are divided into two domains, called Bacteria and Archaea, originally categorized as Eubacteria and Archaebacteria.8 However both terms, Eubacteria and Bacteria are still being used in microbiology.
Eubacteria probe recognizes most bacteria as it is complementary to a portion
of 16S rRNA found in almost all bacteria.9,10
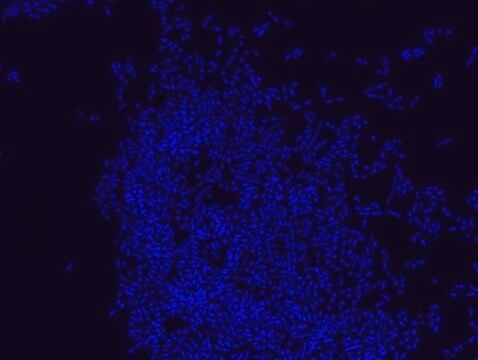
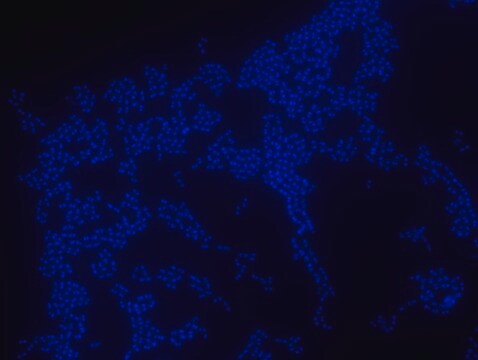
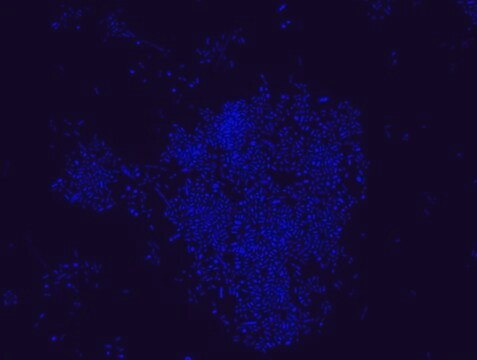
FISH technique was successfully used to identify different bacteria with the universal bacterial probe in various samples such as, pure culture (as described in the figure legends), blood cultures10,11, periapical tooth lesions12, saliva13, biofilms from voice prostheses14, subgingival biofilm15, aortic wall tissue16, buccal epithelial cells, pure culture and cell culture17, intestine tissue embedded in paraffin18, necrotizing fasciitis and pure culture19, colon sections embedded in paraffin20,21, cancer tissues22,23, environmental samples24 and gut of the medicinal leech25. The probe can also be used for combined technique of FISH and Flow cytometric analysis. 9,26,27
It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative (MBD0034/35) control probes, that accompany the specific probe of interest.
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems6) and plants7.
Prokaryotic single cell life forms are divided into two domains, called Bacteria and Archaea, originally categorized as Eubacteria and Archaebacteria.8 However both terms, Eubacteria and Bacteria are still being used in microbiology.
Eubacteria probe recognizes most bacteria as it is complementary to a portion
of 16S rRNA found in almost all bacteria.9,10
FISH technique was successfully used to identify different bacteria with the universal bacterial probe in various samples such as, pure culture (as described in the figure legends), blood cultures10,11, periapical tooth lesions12, saliva13, biofilms from voice prostheses14, subgingival biofilm15, aortic wall tissue16, buccal epithelial cells, pure culture and cell culture17, intestine tissue embedded in paraffin18, necrotizing fasciitis and pure culture19, colon sections embedded in paraffin20,21, cancer tissues22,23, environmental samples24 and gut of the medicinal leech25. The probe can also be used for combined technique of FISH and Flow cytometric analysis. 9,26,27
It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative (MBD0034/35) control probes, that accompany the specific probe of interest.
Application
Probe for fluorescence in situ hybridization (FISH), recognizes Eubacteria cells.
It is recommended to use 20% Formamide for hybridization buffer.
It is recommended to use 20% Formamide for hybridization buffer.
Features and Benefits
- Visualize, identify and isolate bacteria cells.
- Observe native bacteria cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of bacteria cells in mixed microorganism population.
- Specific, sensitive and robust identification even when bacteria are in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on bacteria morphology and allows to study biofilm architecture.
- Identify various bacteria in environmental and clinical samples such as, formalin-fixed paraffin-embedded (FFPE) samples, blood cultures, saliva and more.
- The ability to detect bacteria in its natural habitat is an essential tool for studying host-microbiome interaction.
Disclaimer
Unless otherwise stated in our catalog our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
R I Amann et al.
Applied and environmental microbiology, 56(6), 1919-1925 (1990-06-01)
Fluorescent oligonucleotide hybridization probes were used to label bacterial cells for analysis by flow cytometry. The probes, complementary to short sequence elements within the 16S rRNA common to phylogenetically coherent assemblages of microorganisms, were labeled with tetramethylrhodamine and hybridized to
C R Woese et al.
Proceedings of the National Academy of Sciences of the United States of America, 87(12), 4576-4579 (1990-06-01)
Molecular structures and sequences are generally more revealing of evolutionary relationships than are classical phenotypes (particularly so among microorganisms). Consequently, the basis for the definition of taxa has progressively shifted from the organismal to the cellular to the molecular level.
Sven Poppert et al.
Journal of medical microbiology, 59(Pt 1), 65-68 (2009-10-03)
This study evaluated fluorescence in situ hybridization (FISH) for rapid identification of Staphylococcus aureus and coagulase-negative staphylococci (CoNS) directly from blood cultures. Initially, 360 blood cultures containing Gram-positive cocci were investigated by a previously described microwave-FISH procedure: 44/49 (89.8 %)
Leore T Geller et al.
Science (New York, N.Y.), 357(6356), 1156-1160 (2017-09-16)
Growing evidence suggests that microbes can influence the efficacy of cancer therapies. By studying colon cancer models, we found that bacteria can metabolize the chemotherapeutic drug gemcitabine (2',2'-difluorodeoxycytidine) into its inactive form, 2',2'-difluorodeoxyuridine. Metabolism was dependent on the expression of
Cathleen Schlundt et al.
Molecular ecology resources, 20(3), 620-634 (2019-11-30)
Plastic marine debris (PMD) affects spatial scales of life from microbes to whales. However, understanding interactions between plastic and microbes in the "Plastisphere"-the thin layer of life on the surface of PMD-has been technology-limited. Research into microbe-microbe and microbe-substrate interactions
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service