MBD0059
Alphaproteobacteria FISH probe-Cy3
Probe for fluorescence in situ hybridization (FISH), 20 μM in water
Sinónimos:
ALF968
Iniciar sesiónpara Ver la Fijación de precios por contrato y de la organización
About This Item
Código UNSPSC:
12352200
NACRES:
NA.55
Productos recomendados
Nivel de calidad
concentración
20 μM in water
fluorescencia
λex 550 nm; λem 570 nm (Cy3)
Condiciones de envío
dry ice
temp. de almacenamiento
-10 to -25°C
Descripción general
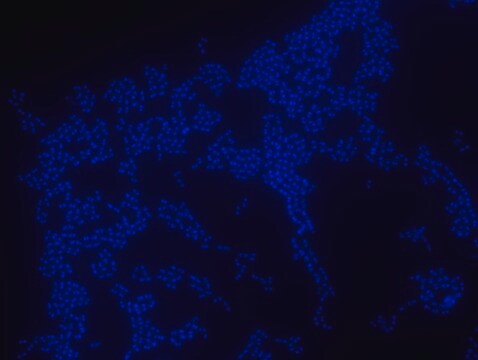
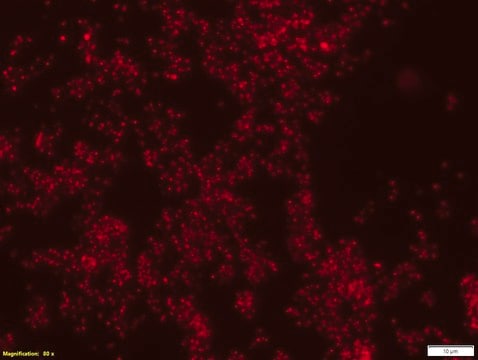
FISH (Fluorescent in Situ Hybridization) technique is based on the hybridization of fluorescently labeled oligonucleotide probes to a specific complementary DNA or RNA sequence in whole and intact cells. Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples. FISH can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood and other tissues), microbial ecology (solid biofilms and aquatic systems) and plants. Alphaproteobacteria FISH probe – Cy3 (MBD0059) specifically recognizes Alphaproteobacteria cells. Alphaproteobacteria is the second largest class of bacteria within the gram-negative phylum Proteobacteria, with a high level of functional diversity among its eight major orders: Caulobacterales, Rhizobiales, Rhodobacterales, Pelagibacterales, Sphingomonadales, Rhodospirillales, Holosporales and Rickettsiales. Mitochondria are thought to have evolved from Alphaproteobacteria. Between 20-50% of bacterioplankton cells in the ocean belong to the Alphaproteobacteria class, and Alphaproteobacteria is also abundant in soil and freshwater environments. Rhizobiales is a soil-based plant symbiont Alphaproteobacteria widely studied in agricultural research and Rhodobacterales is an example of a purple photosynthetic Alphaproteobacteria bacteria. Certain genera of Alphaproteobacteria, such as Brucella, are common human pathogens. One species of Alphaproteobacteria, Paracoccus yeei, was correlated to opportunistic infections in humans, including peritonitis,myocarditis, and dermatologic lesions. Interestingly, several types of Alphaproteobacteria have been shown to divide asymmetrically, a trait that may contribute to this class’s heterogeneity and adaptability to a wide variety of microbiome environments. The FISH probe has been used to recognize many Alphaproteobacteria species, such as Magnetospirillum gryphiswaldense, a spiral-shaped magnetotactic bacteria first isolated from river sediment in 1990. In addition, the FISH probe has been used to detect Alphaproteobacteria from a wide variety of environmental microbial samples, including bacterial populations isolated from snow, wastewater treatment plants, lichen and sphagnum moss samples, seawater, and freshwater sediments.
Aplicación
Probe for fluorescence in situ hybridization (FISH), recognizes Alphaproteobacteria cells (except for Rickettsiales)
Características y beneficios
- Visualize, identify and isolate Alphaproteobacteria cells.
- Observe native Alphaproteobacteria cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of Alphaproteobacteria in bacterial mixed populations.
- Specific, sensitive and robust identification even when Alphaproteobacteria is in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on Alphaproteobacteria morphology
Código de clase de almacenamiento
12 - Non Combustible Liquids
Clase de riesgo para el agua (WGK)
WGK 1
Punto de inflamabilidad (°F)
Not applicable
Punto de inflamabilidad (°C)
Not applicable
Elija entre una de las versiones más recientes:
Certificados de análisis (COA)
Lot/Batch Number
¿No ve la versión correcta?
Si necesita una versión concreta, puede buscar un certificado específico por el número de lote.
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Genome streamlining in a cosmopolitan oceanic bacterium
Science, 309(5738), 1242-1245 (2005)
In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria
Cardinale, et al.
FEMS Microbiology Ecology, 66(1), 63-71 (2008)
Developmental and environmental regulatory pathways in alpha-proteobacteria
Frontiers in Bioscience (Landmark Edition), 17(5), 1695-1714 (2012)
An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins
eLife, 8 (2019)
Paracoccus yeeii sp. nov. (formerly CDC group EO-2), a novel bacterial species associated with human infection
Maryam I Daneshvar
Journal of Clinical Microbiology, 41(3), 1289-1294 (2003)
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico