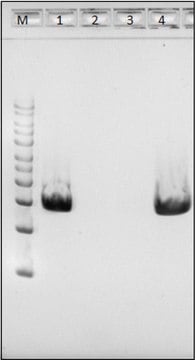
MBD6000
5R-Plex kit
Ultra-sensitive 16S NGS assay for degraded and low biomass DNA
Se connecterpour consulter vos tarifs contractuels et ceux de votre entreprise/organisme
About This Item
Code UNSPSC :
41106302
Nomenclature NACRES :
NA.84
Produits recommandés
Niveau de qualité
Utilisation
Preparing 96 samples for sequencing
Technique(s)
DNA amplification: suitable
DNA sequencing: suitable
Conditions d'expédition
dry ice
Température de stockage
-10 to -25°C
Description générale
The 16S ribosomal RNA gene (16S rRNA) is a common bacterial marker, used as a target for microbial community profiling in microbiome metagenomic studies. The gene is composed of nine variable regions (V1-V9) interspersed between conserved regions. The most common 16S rRNA NGS assays target one or two regions using a single set of primers (e.g., V3-V4). However, this approach usually results in a poor bacterial detection and classification rate when the DNA input is of low-quality or when applying it on extremely low biomass samples. The 5R-PLEX kit offers a complete solution to challenging biological samples where the standard 16S sequencing protocols fail to perform. Sample types include:
- Formalin-Fixed, Paraffin-Embedded (FFPE) tissue
- Cancerous tumor tissue
- Degraded or damaged DNA
- Low biomass samples
- Fossil-derived and ancient DNA
Caractéristiques et avantages
- The 5R-PLEX Kit provides high profile sequencing from degraded, damaged or low biomass samples
- Multiplexed NGS assay targets 5 variable regions of the 16S rRNA gene in a single primer pool
- Includes positive control for trouble shooting experiments.
- The kit can detect as low as 1pg of highly degraded bacterial DNA with an average fragment size of 260bp.
- Kit includes low bioburden reagents
- Includes exclusive access to M-CAMP™ platform featuring an innovative algorithm designed to reconstruct a single coherent microbial profiling and perform comprehensive analysis.
Composants
- 5R-PLEX PCR1 Primer mix- 25 μL
- 5R-PLEX PCR2 Forward Primer mix- 25 μL
- 5R-PLEX index plate 96-plate
- Water, microbial DNA-free 10X- 1.5 mL
- HF DNA Polymerase- 0.1 mL
- 5X HF buffer 2 mL
- dNTP′s- 0.2 mL
- Elution Buffer (EB), microbial DNA-free- 8 mL
- 5R-PLEX Positive Control (10 ng/μL)- 30 μL
Stockage et stabilité
The shelf life of all reagents provided in the kit is 12 months when stored properly. Store all components at -20 °C.
Autres remarques
Kit component details can be found in the User guide.
Informations légales
M-Camp is a trademark of Merck KGaA, Darmstadt, Germany
Mentions de danger
Conseils de prudence
Classification des risques
Aquatic Chronic 3
Code de la classe de stockage
10 - Combustible liquids
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Nicole M Davis et al.
Microbiome, 6(1), 226-226 (2018-12-19)
The accuracy of microbial community surveys based on marker-gene and metagenomic sequencing (MGS) suffers from the presence of contaminants-DNA sequences not truly present in the sample. Contaminants come from various sources, including reagents. Appropriate laboratory practices can reduce contamination, but
Jacob T Nearing et al.
Microbiome, 9(1), 113-113 (2021-05-20)
Advances in DNA sequencing technology have vastly improved the ability of researchers to explore the microbial inhabitants of the human body. Unfortunately, while these studies have uncovered the importance of these microbial communities to our health, they often do not
Deborah Nejman et al.
Science (New York, N.Y.), 368(6494), 973-980 (2020-05-30)
Bacteria were first detected in human tumors more than 100 years ago, but the characterization of the tumor microbiome has remained challenging because of its low biomass. We undertook a comprehensive analysis of the tumor microbiome, studying 1526 tumors and
Amnon Amir et al.
Nucleic acids research, 41(22), e205-e205 (2013-11-12)
The emergence of massively parallel sequencing technology has revolutionized microbial profiling, allowing the unprecedented comparison of microbial diversity across time and space in a wide range of host-associated and environmental ecosystems. Although the high-throughput nature of such methods enables the
Garold Fuks et al.
Microbiome, 6(1), 17-17 (2018-01-28)
Most of our knowledge about the remarkable microbial diversity on Earth comes from sequencing the 16S rRNA gene. The use of next-generation sequencing methods has increased sample number and sequencing depth, but the read length of the most widely used
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique