293768
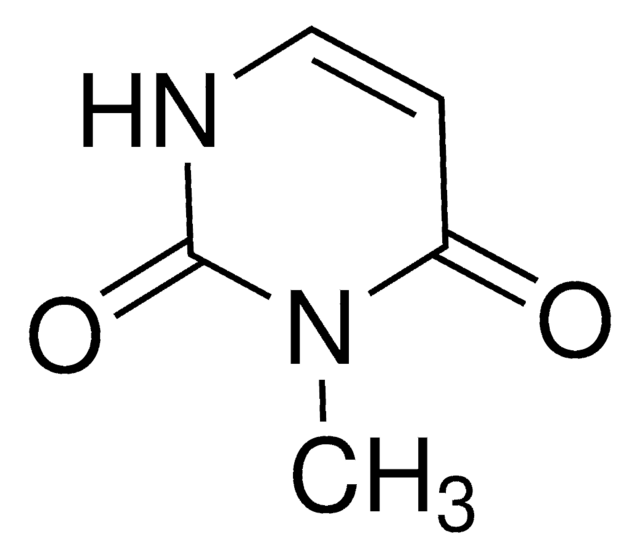
1-Methyluracil
99%
Synonym(s):
1-Methyl-2,4(1H,3H)-pyrimidinedione
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
Empirical Formula (Hill Notation):
C5H6N2O2
CAS Number:
Molecular Weight:
126.11
MDL number:
UNSPSC Code:
12352100
PubChem Substance ID:
NACRES:
NA.22
Recommended Products
Quality Level
Assay
99%
mp
236-238 °C (lit.)
solubility
1 M NaOH: soluble 50 mg/mL, clear, colorless
SMILES string
CN1C=CC(=O)NC1=O
InChI
1S/C5H6N2O2/c1-7-3-2-4(8)6-5(7)9/h2-3H,1H3,(H,6,8,9)
InChI key
XBCXJKGHPABGSD-UHFFFAOYSA-N
General description
1-Methyluracil is of special importance in biochemistry, since uracil attaches ribose in ribonucleic acid (RNA) just precisely at the N1 atom. H-bond complex formation between 1-methyluracil and glycine has been investigated by theoretical calculations and FT-IR spectroscopy in Ar matrices. It forms 1:1 complexes with 9-ethyl-8-bromo-2,6-diaminopurine and the complex structure has been determined by three-dimensional X-ray diffraction methods.
Signal Word
Warning
Hazard Statements
Precautionary Statements
Hazard Classifications
Carc. 2
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
R K McMullan et al.
Acta crystallographica. Section B, Structural science, 45 ( Pt 3), 270-276 (1989-06-01)
The crystal structure of 1-methylpyrimidine-2,4-dione (1-methyluracil, C5H6N2O2) has been determined at 15, 60 and 123 K from neutron diffraction data. Molecules lie in the eightfold special positions (symmetry m) of space group Ibam, with a = 13.213 (2), b =
V I Poltev et al.
Molekuliarnaia biologiia, 29(2), 365-375 (1995-03-01)
Monte Carlo simulation of hydration of keto and enol tautomers of 9-methylguanine (G) and 1-methyluracil (U) has been performed in relation to a possible role of tautomer transitions of DNA bases in mutagenesis. The comparison of the simulation results with
Bram Boeckx et al.
The journal of physical chemistry. B, 116(39), 11890-11898 (2012-09-12)
The H-bond complex formation between 1-methyluracil and glycine has been investigated by theoretical calculations and the most stable complex configurations have been identified by FT-IR spectroscopy in Ar matrices. The importance of this H-bonding system is huge since all DNA
V I Poltev et al.
Journal of biomolecular structure & dynamics, 9(1), 101-111 (1991-08-01)
A number of nucleic acid base pairs and complexes between the bases and the amide group of acrylamide have been studied experimentally by using mass spectrometry and theoretically by the method of atom-atom potential function calculations. It has been found
Base-pairing configurations between purines and pyrimidines in the solid state. V. Crystal and molecular structure of two 1:1 hydrogen-bonded complexes, 1-methyluracil: 9-ethyl-8-bromo-2,6-diaminopurine and 1-ethylthymine: 9-ethyl-8-bromo-2,6--diaminopurine.
G Simundza et al.
Journal of molecular biology, 48(2), 263-278 (1970-03-14)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service





![6-Methylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione](/deepweb/assets/sigmaaldrich/product/structures/393/943/c932f315-dd4b-4939-aea6-646238005e48/640/c932f315-dd4b-4939-aea6-646238005e48.png)


