All Photos(1)
About This Item
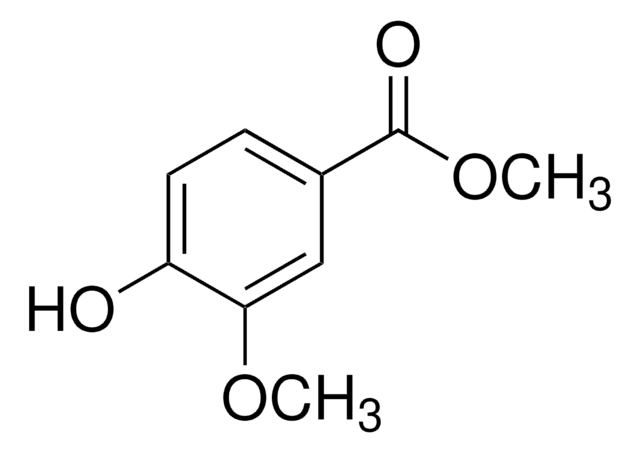
Linear Formula:
ClC6H4CO2H
CAS Number:
Molecular Weight:
156.57
Beilstein:
907218
EC Number:
MDL number:
UNSPSC Code:
12352100
Recommended Products
mp
153-157 °C (lit.)
SMILES string
OC(=O)c1cccc(Cl)c1
InChI
1S/C7H5ClO2/c8-6-3-1-2-5(4-6)7(9)10/h1-4H,(H,9,10)
InChI key
LULAYUGMBFYYEX-UHFFFAOYSA-N
Looking for similar products? Visit Product Comparison Guide
replaced by
Product No.
Description
Pricing
Signal Word
Warning
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2 - Skin Irrit. 2
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Alfredo Gallego et al.
World journal of microbiology & biotechnology, 28(3), 1245-1252 (2012-07-19)
An indigenous strain of Pseudomonas putida capable of degrading 3-chlorobenzoic acid as the sole carbon source was isolated from the Riachuelo, a polluted river in Buenos Aires. Aerobic biodegradation assays were performed using a 2-l microfermentor. Biodegradation was evaluated by
Caroline Laemmli et al.
Archives of microbiology, 181(2), 112-121 (2003-12-17)
Ralstonia eutropha JMP134 possesses two sets of similar genes for degradation of chloroaromatic compounds, tfdCDEFB (in short: tfdI cluster) and tfdDII CII EII FII BII (tfdII cluster). The significance of two sets of tfd genes for the organism has long
Hee-Sung Bae et al.
Chemosphere, 55(1), 93-100 (2004-01-15)
An anaerobic continuous-flow fixed-bed column reactor capable of degrading 3-chlorobenzoate (3-CBA) under denitrifying conditions was established, and its rate reached 2.26 mM d(-1). The denitrifying population completely degraded 3-CBA when supplied at 0.1-0.54 mM, but its activity was partly suppressed
Sudip K Samanta et al.
Molecular microbiology, 55(4), 1151-1159 (2005-02-03)
Rhodopseudomonas palustris strain RCB100 degrades 3-chlorobenzoate (3-CBA) anaerobically. We purified from this strain a coenzyme A ligase that is active with 3-CBA and determined its N-terminal amino acid sequence to be identical to that of a cyclohexanecarboxylate-CoA ligase encoded by
V P Jayachandran et al.
Journal of industrial microbiology & biotechnology, 36(2), 219-227 (2008-10-23)
The compatibility and efficiency of two ortho-cleavage pathway-following pseudomonads viz. the 3-chlorobenzoate (3-CBA)-degrader, Pseudomonas aeruginosa 3mT (3mT) and the phenol-degrader, P. stutzeri SPC-2 (SPC-2) in a mixed culture for the degradation of these substrates singly and simultaneously in mixtures was
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service