P9474
Pyruvate Decarboxylase from baker′s yeast (S. cerevisiae)
ammonium sulfate suspension, 5.0-20.0 units/mg protein (biuret)
Synonym(s):
2-Oxo-acid carboxy-lyase
Sign Into View Organizational & Contract Pricing
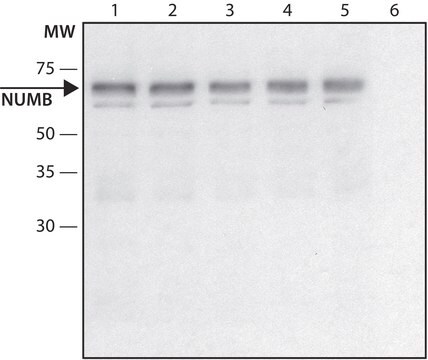
All Photos(1)
About This Item
CAS Number:
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.54
Recommended Products
form
ammonium sulfate suspension
specific activity
5.0-20.0 units/mg protein (biuret)
storage temp.
2-8°C
Looking for similar products? Visit Product Comparison Guide
General description
Pyruvate decarboxylase (PDC) usually appear in plant seeds at the time of germination, especially when the plant embryo is totally covered by an oxygen-impermeable testa.
Application
Pyruvate Decarboxylase from baker′s yeast (S. cerevisiae) has been used to evaluate the power of systematic identification of meaningful metabolic enzyme regulation (SIMMER) for finding unknown yeast regulatory interactions.
Pyruvate decarboxylase (PDC) is used to study residues involved in thiamine pyrophosphate (TPP) binding. It is used to study the regulation of fermentation pathways in plant species.
Biochem/physiol Actions
Pyruvate decarboxylase (PDC) is a homotetrameric enzyme that catalyses the decarboxylation of pyruvic acid to acetaldehyde and carbon dioxide in the cytoplasm. Pyruvate decarboxylase depends on cofactors thiamine pyrophosphate (TPP) and magnesium. PDC contains a β-α-β structure, yielding parallel β-sheets.
Pyruvate decarboxylase (PDC) actively participates in the anaerobic metabolism of several bacteria, yeast and plant seeds.
Unit Definition
One unit will convert 1.0 μmole of pyruvate to acetaldehyde per min at pH 6.0 at 25 °C.
Physical form
Suspension in 3.2 M (NH4)2SO4 pH 6.5, stabilized with 5% glycerol, 5 mM potassium phosphate, 1 mM magnesium acetate, 0.5 mM EDTA, and 25 μM cocarboxylase.
Preparation Note
Isolated without the use of heavy metals.
Storage Class Code
10 - Combustible liquids
WGK
WGK 2
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Clara C Posthuma et al.
Applied and environmental microbiology, 68(2), 831-837 (2002-02-02)
Purification of xylulose 5-phosphate phosphoketolase (XpkA), the central enzyme of the phosphoketolase pathway (PKP) in lactic acid bacteria, and cloning and sequence analysis of the encoding gene, xpkA, from Lactobacillus pentosus MD363 are described. xpkA encodes a 788-amino-acid protein with
Systems-level analysis of mechanisms regulating yeast metabolic flux
Hackett S R, et al.
Science, 354(6311), aaf2786-aaf2786 (2016)
Bijayalaxmi Mohanty et al.
Annals of botany, 91 Spec No, 291-300 (2003-01-02)
Submergence tolerance of 13 doubled haploid lines of rice and their parents (submergence tolerant FR13A and submergence intolerant CT6241) was assessed using 2-week-old seedlings. Plants were scored for leaf senescence and percentage of seedlings that survived up to 15 d
F Dyda et al.
Biochemistry, 32(24), 6165-6170 (1993-06-22)
The crystal structure of brewers' yeast pyruvate decarboxylase, a thiamin diphosphate dependent alpha-keto acid decarboxylase, has been determined to 2.4-A resolution. The homotetrameric assembly contains two dimers, exhibiting strong intermonomer interactions within each dimer but more limited ones between dimers.
Substrate activation behaviour of pyruvate decarboxylase from Pisum sativum cv. Miko
Dietrich A and Konig S
Febs Letters, 400(1), 42-44 (1997)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service