SAMMS2HYG
MS2-P65-HSF1-Hygromycin SAM CRISPRa Helper Construct 2 Plasmid DNA
Se connecterpour consulter vos tarifs contractuels et ceux de votre entreprise/organisme
About This Item
Code UNSPSC :
12352200
Nomenclature NACRES :
NA.51
Produits recommandés
Conditionnement
vial of 20 μL
Concentration
500 ng/μL in TE buffer; DNA (10μg of plasmid DNA)
Application(s)
CRISPR
Conditions d'expédition
dry ice
Température de stockage
−20°C
Description générale
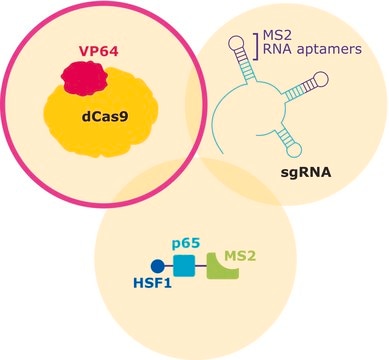
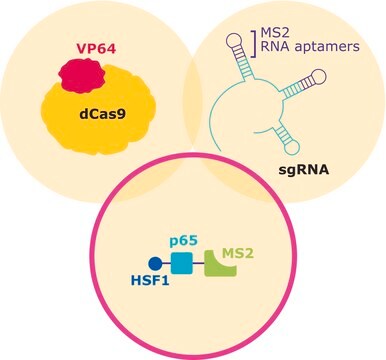
This product is a lentiviral plasmid that utilizes the EF1 alpha promoter to drive expression of MS2-P65-HSF1 and hygromycin resistance cassette linked by a 2A peptide (EF1a-MS2-P65-HSF1-2A-Hygromycin) allowing for easy selection following successful transfection or transduction. Use Sigma′s lentiviral MS2-P65-HSF1 plasmid for generation of lentiviral particles and efficient production of stable cell lines expressing MS2-P65-HSF1 for CRISPR based transcipritonal activation and genome wide gain of function screening. Sigma′s lenti-MS2-P65-HSF1 plasmid is one part of a three part CRISPR system with individual dCas9-VP64, MS2-p65-HSF1 and gRNA expression vectors.
To order gRNA in any format click here
To order gRNA in any format click here
Application
Functional Genomics/Target Validation
- Unbiased forward genetic screening
- Strong transcriptional activation in multiple cell lines
- Manufacture of MS2-p65-HSF1 expressing Lentiviral Particles
Caractéristiques et avantages
- Highly specific and highly active
- Sequence verified high purity plasmid DNA
- Activates genes through transcriptional activation rather than cDNA based overexpression
Learn more about SAM CRISPR Activators at SigmaAldrich.com/CRISPRa
Principe
CRISPR/Cas systems are employed by bacteria and archaea as a defense against invading viruses and plasmids. Recently, the type II CRISPR/Cas system from the bacterium Streptococcus pyogenes has been engineered to function in eukaryotic systems using two molecular components: a single Cas9 protein and a non-coding guide RNA (gRNA). The Cas9 endonuclease can be rendered inactive (dCas9) with mutations to the two protein domains, RuvC and HnH (D10A and H840A respectively), which are responsible for nuclease activity. The nuclease deficient protein can then be fused with the transcriptional activator VP64 and used in conjunction with a guide RNA modified with MS2 RNA aptamers that function to recruit the additional transcriptional coactivators p65 and HSF1. The assembled SAM complex is then used as a cargo delivery system to target gene promoters, enabling site-specific transcriptional activation of the gene of interest
Informations légales
Code de la classe de stockage
12 - Non Combustible Liquids
Classe de danger pour l'eau (WGK)
WGK 2
Point d'éclair (°F)
Not applicable
Point d'éclair (°C)
Not applicable
Certificats d'analyse (COA)
Recherchez un Certificats d'analyse (COA) en saisissant le numéro de lot du produit. Les numéros de lot figurent sur l'étiquette du produit après les mots "Lot" ou "Batch".
Déjà en possession de ce produit ?
Retrouvez la documentation relative aux produits que vous avez récemment achetés dans la Bibliothèque de documents.
Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex
Konermann S, et al
Nature, 517, 83-588 (2015)
Genome-scale CRISPR-Cas9 Knockout and Transcriptional Activation Screening.
Joung, S. et al.
Nature Protocols, 12, 828-863 (2017)
Julia Joung et al.
Nature protocols, 12(4), 828-863 (2017-03-24)
Forward genetic screens are powerful tools for the unbiased discovery and functional characterization of specific genetic elements associated with a phenotype of interest. Recently, the RNA-guided endonuclease Cas9 from the microbial CRISPR (clustered regularly interspaced short palindromic repeats) immune system
Notre équipe de scientifiques dispose d'une expérience dans tous les secteurs de la recherche, notamment en sciences de la vie, science des matériaux, synthèse chimique, chromatographie, analyse et dans de nombreux autres domaines..
Contacter notre Service technique