D6319
Anti-DCP2 (C-terminal) antibody produced in rabbit
~1 mg/mL, affinity isolated antibody, buffered aqueous solution
Synonym(s):
Anti-DCP2 decapping enzyme homolog (S. cerevisiae), Anti-NUDT20, Anti-Nucleleoside diphosphate-linked moiety X motif 20, Anti-Nudix motif 20
About This Item
Recommended Products
biological source
rabbit
conjugate
unconjugated
antibody form
affinity isolated antibody
antibody product type
primary antibodies
clone
polyclonal
form
buffered aqueous solution
mol wt
antigen ~50 kDa
species reactivity
rat, mouse, human
concentration
~1 mg/mL
technique(s)
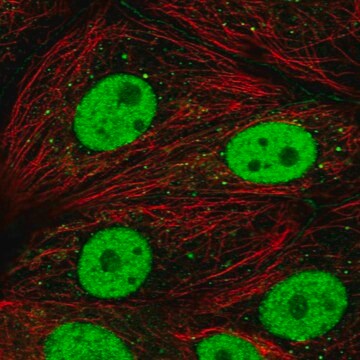
indirect immunofluorescence: 2-5 μg/mL using paraformaldehyde-fixed NIH-3T3 celle
indirect immunofluorescence: suitable
western blot: 2-4 μg/mL using lysaes of K-562 and Rat1 cells
UniProt accession no.
shipped in
dry ice
storage temp.
−20°C
target post-translational modification
unmodified
Gene Information
human ... DCP2(167227)
mouse ... Dcp2(20640)
rat ... Dcp2(291604)
General description
Immunogen
Application
Biochem/physiol Actions
Target description
Physical form
Disclaimer
Not finding the right product?
Try our Product Selector Tool.
Storage Class Code
10 - Combustible liquids
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service