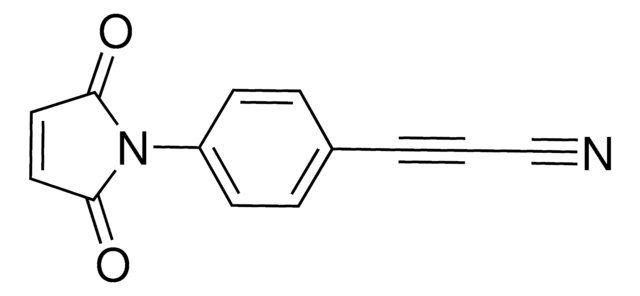
760668
Dibenzocyclooctyne-maleimide
for Copper-free Click Chemistry
Sinónimos:
DBCO-maleimide
About This Item
Productos recomendados
Quality Level
form
solid
reaction suitability
reaction type: click chemistry
reagent type: cross-linking reagent
functional group
maleimide
storage temp.
−20°C
SMILES string
O=C(N1CCC(NCCC(N2CC3=C(C=CC=C3)C#CC4=C2C=CC=C4)=O)=O)C=CC1=O
InChI
1S/C25H21N3O4/c29-22(14-16-27-23(30)11-12-24(27)31)26-15-13-25(32)28-17-20-7-2-1-5-18(20)9-10-19-6-3-4-8-21(19)28/h1-8,11-12H,13-17H2,(H,26,29)
InChI key
NHFQNAGPXIVKND-UHFFFAOYSA-N
Categorías relacionadas
Application
Applications Include:
- Protein-peptide conjugates
- Antibody-enzyme or antibody-drug conjugates
- Protein or peptide-oligonucleotide conjugates
- Surface modification
Elija entre una de las versiones más recientes:
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Artículos
Copper-free click chemistry is an alternative approach to click chemistry that proceeds at a lower activation barrier and is free of cytotoxic transition metal catalysts.
Copper-free click chemistry is an alternative approach to click chemistry that proceeds at a lower activation barrier and is free of cytotoxic transition metal catalysts.
Copper-free click chemistry is an alternative approach to click chemistry that proceeds at a lower activation barrier and is free of cytotoxic transition metal catalysts.
Copper-free click chemistry is an alternative approach to click chemistry that proceeds at a lower activation barrier and is free of cytotoxic transition metal catalysts.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico


![[1,1′-bis(difenilfosfino)ferroceno]dicloropaladio(II), complejo con diclorometano](/deepweb/assets/sigmaaldrich/product/structures/825/986/4317978b-1256-4c82-ab74-6a6a3ef948b1/640/4317978b-1256-4c82-ab74-6a6a3ef948b1.png)


![[1,1′-Bis(diphenylphosphino)ferrocene]dichloropalladium(II)](/deepweb/assets/sigmaaldrich/product/structures/130/734/8846aa26-1858-458a-998d-8c306c13bf0f/640/8846aa26-1858-458a-998d-8c306c13bf0f.png)


![(1R,8S,9s)-Bicyclo[6.1.0]non-4-yn-9-ylmethyl N-succinimidyl carbonate for Copper-free Click Chemistry](/deepweb/assets/sigmaaldrich/product/structures/969/022/d6776082-2f7a-47c7-bcd4-3830dac0fb7d/640/d6776082-2f7a-47c7-bcd4-3830dac0fb7d.png)


![N-[(1R,8S,9s)-Bicyclo[6.1.0]non-4-yn-9-ylmethyloxycarbonyl]-1,8-diamino-3,6-dioxaoctane for Copper-free Click Chemistry](/deepweb/assets/sigmaaldrich/product/structures/294/853/c5e47d84-5aee-4797-aa24-604f291171cc/640/c5e47d84-5aee-4797-aa24-604f291171cc.png)
![(1R,8S,9s)-Bicyclo[6.1.0]non-4-yn-9-ylmethanol for Copper-free Click Chemistry](/deepweb/assets/sigmaaldrich/product/structures/171/632/0556139a-2db5-4678-a6ec-a26a693fd574/640/0556139a-2db5-4678-a6ec-a26a693fd574.png)