SML1248
KT182
≥98% (HPLC)
Sinónimos:
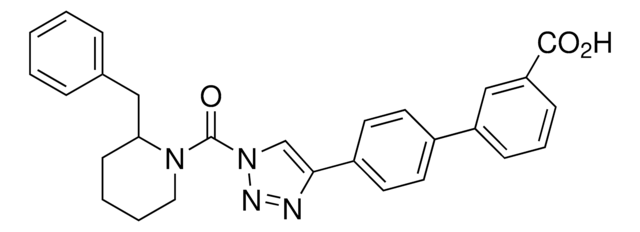
[4-[3′-(Hydroxymethyl)[1,1′-biphenyl]-4-yl]-1H-1,2,3-triazol-1-yl](2-phenyl-1-piperidinyl)-methanone
About This Item
Productos recomendados
Quality Level
assay
≥98% (HPLC)
form
powder
color
white to beige
solubility
DMSO: 10 mg/mL, clear
storage temp.
2-8°C
SMILES string
O=C(N1N=NC(C2=CC=C(C3=CC=CC(CO)=C3)C=C2)=C1)N4CCCCC4C5=CC=CC=C5
InChI
1S/C27H26N4O2/c32-19-20-7-6-10-24(17-20)21-12-14-22(15-13-21)25-18-31(29-28-25)27(33)30-16-5-4-11-26(30)23-8-2-1-3-9-23/h1-3,6-10,12-15,17-18,26,32H,4-5,11,16,19H2
InChI key
GICNKPZHUCVFNM-UHFFFAOYSA-N
¿Está buscando productos similares? Visita Guía de comparación de productos
Biochem/physiol Actions
signalword
Warning
hcodes
Hazard Classifications
Acute Tox. 4 Oral - Aquatic Chronic 4
Storage Class
11 - Combustible Solids
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Contenido relacionado
The aim of the Cravatt research group is to understand the roles that mammalian enzymes play in physiological and pathological processes and to use this knowledge to identify novel therapeutic targets for the treatment of human disease. To achieve these goals, they develop and apply new technologies that bridge the fields of chemistry and biology, ascribing to the philosophy that the most significant biomedical problems require creative multidisciplinary approaches for their solution. The group's technological innovations address fundamental challenges in systems biology that are beyond the scope of contemporary methods. For instance, enzymes are tightly regulated by post-translational events in vivo, meaning that their activity may not correlate with expression as measured by standard genomic and proteomic approaches. Considering that it is an enzyme's activity, rather than abundance that ultimately dictates its role in cell physiology and pathology, the Cravatt group has introduced a set of proteomic technologies that directly measures this parameter. These activity-based protein profiling (ABPP) methods exploit the power of chemistry to engender new tools and assays for the global analysis of enzyme activities. The enzyme activity profiles generated by ABPP constitute unique molecular portraits of cells and tissues that illuminate how metabolic and signaling networks are regulated in vivo. Additionally, by evaluating enzymes based on functional properties rather than mere abundance, ABPP acquires high-content proteomic information that is enriched in novel markers and targets for the diagnosis and treatment of human disease.
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico