930431
SuTEx1-alkyne
≥95%
Synonym(s):
N-2-propyn-1-yl-4-(1H-1,2,4-triazol-1-ylsulfonyl)-benzamide
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
Empirical Formula (Hill Notation):
C12H10N4O3S
CAS Number:
Molecular Weight:
290.30
UNSPSC Code:
12352101
NACRES:
NA.21
Recommended Products
description
Application: Chemoproteomics
Quality Level
Assay
≥95%
form
powder
mp
155 °C
storage temp.
−20°C
SMILES string
O=C(C1=CC=C(S(N2N=C([H])N=C2)(=O)=O)C=C1)NCC#C
Application
SuTEx1-alkyne is a probe that uses sulfur-triazole exchange chemistry to label tyrosines. A method was developed using cysteine-reactive compounds including this one to allow for unbiased analysis of proteomic data in quantitative applications (Zanon et al. 2021). The method uses light or heavy labelling with the isotopically labelled desthiobiotin azide (isoDTB) tag for mass spectrometry analysis (Zanon et al. 2020). Analysis then uses the isotopic tandem orthogonal proteolysis activity-based protein profiling (isoTOP-ABPP) workflow (Weerapana et al. 2010, Backus et al. 2016)
Other Notes
Profiling the proteome-wide selectivity of diverse electrophiles
A quantitative thiol reactivity profiling platform to analyze redox and electrophile reactive cysteine proteomes
Ethynylation of Cysteine Residues: From Peptides to Proteins in Vitro and in Living Cells
A Chemoproteomic Platform To Assess Bioactivation Potential of Drugs
Inhibition of Zinc-Dependent Histone Deacetylases with a Chemically Triggered Electrophile
Reversibility of Covalent Electrophile-Protein Adducts and Chemical Toxicity
A quantitative thiol reactivity profiling platform to analyze redox and electrophile reactive cysteine proteomes
Ethynylation of Cysteine Residues: From Peptides to Proteins in Vitro and in Living Cells
A Chemoproteomic Platform To Assess Bioactivation Potential of Drugs
Inhibition of Zinc-Dependent Histone Deacetylases with a Chemically Triggered Electrophile
Reversibility of Covalent Electrophile-Protein Adducts and Chemical Toxicity
related product
Product No.
Description
Pricing
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Lot/Batch Number
Sorry, we don't have COAs for this product available online at this time.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Profiling the proteome-wide selectivity of diverse electrophiles
Patrick R. A. Zanon ,Fengchao Yu, et al
ChemRxiv : the preprint server for chemistry (2021)
Rui Sun et al.
Chemical research in toxicology, 30(10), 1797-1803 (2017-09-30)
Reactive metabolites (RM) formed from bioactivation of drugs can covalently modify liver proteins and cause mechanism-based inactivation of major cytochrome P450 (CYP450) enzymes. Risk of bioactivation of a test compound is routinely examined as part of lead optimization efforts in
Ping Yu et al.
Nucleosides, nucleotides & nucleic acids, 40(7), 754-766 (2021-06-29)
We report herein comprehensive investigations of alkylation/sulfur exchange reactions of sulfur-containing substrates including nucleosides such as s2U, m5s2U, s4U, s2A and s2T-incorporated DNA enable by comprehensive screenings of the reagents (2a-2h). It has been proven that iodoacetamide (2a) displays the
Yide He et al.
Talanta, 134, 468-475 (2015-01-27)
In this work, we present a two-step labeling approach for the efficient tagging with lanthanide-containing complexes. For this purpose, derivatization of the cysteine residues with an alkyne group acting as linker was done before the DOTA complex was introduced using
Zarko V Boskovic et al.
ACS chemical biology, 11(7), 1844-1851 (2016-04-12)
Unbiased binding assays involving small-molecule microarrays were used to identify compounds that display unique patterns of selectivity among members of the zinc-dependent histone deacetylase family of enzymes. A novel, hydroxyquinoline-containing compound, BRD4354, was shown to preferentially inhibit activity of HDAC5
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service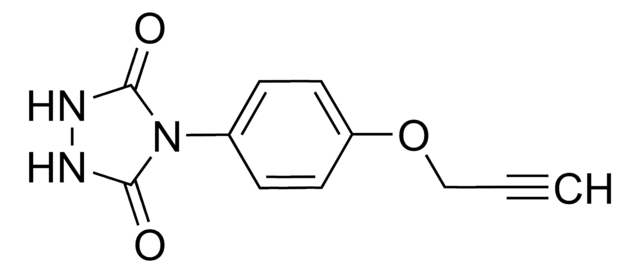


![Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine 97%](/deepweb/assets/sigmaaldrich/product/structures/179/695/86a721c8-2a4c-4e4f-bc36-6276ce7a941f/640/86a721c8-2a4c-4e4f-bc36-6276ce7a941f.png)