D6319
Anti-DCP2 (C-terminal) antibody produced in rabbit
~1 mg/mL, affinity isolated antibody, buffered aqueous solution
别名:
Anti-DCP2 decapping enzyme homolog (S. cerevisiae), Anti-NUDT20, Anti-Nucleleoside diphosphate-linked moiety X motif 20, Anti-Nudix motif 20
About This Item
推荐产品
生物源
rabbit
共軛
unconjugated
抗體表格
affinity isolated antibody
抗體產品種類
primary antibodies
無性繁殖
polyclonal
形狀
buffered aqueous solution
分子量
antigen ~50 kDa
物種活性
rat, mouse, human
濃度
~1 mg/mL
技術
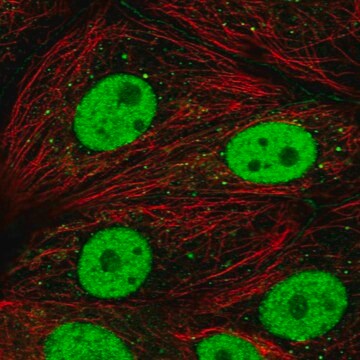
indirect immunofluorescence: 2-5 μg/mL using paraformaldehyde-fixed NIH-3T3 celle
indirect immunofluorescence: suitable
western blot: 2-4 μg/mL using lysaes of K-562 and Rat1 cells
UniProt登錄號
運輸包裝
dry ice
儲存溫度
−20°C
目標翻譯後修改
unmodified
基因資訊
human ... DCP2(167227)
mouse ... Dcp2(20640)
rat ... Dcp2(291604)
一般說明
免疫原
應用
生化/生理作用
標靶描述
外觀
免責聲明
未找到合适的产品?
试试我们的产品选型工具.
儲存類別代碼
10 - Combustible liquids
閃點(°F)
Not applicable
閃點(°C)
Not applicable
個人防護裝備
Eyeshields, Gloves, multi-purpose combination respirator cartridge (US)
我们的科学家团队拥有各种研究领域经验,包括生命科学、材料科学、化学合成、色谱、分析及许多其他领域.
联系技术服务部门