SAB4200609
Monoclonal Anti-diMethyl-Histone H3 (diMe-Lys9) (H3K9me2) antibody produced in mouse
~1.0 mg/mL, clone 5E5-G5, purified immunoglobulin
Synonym(s):
Anti-H3F3A H3.3A H3F3 PP781 H3F3B H3.3B, Anti-HIST1H3A H3FA, Anti-HIST1H3B H3FL, Anti-HIST1H3C H3FC, Anti-HIST1H3D H3FB, Anti-HIST1H3E H3FD, Anti-HIST1H3F H3FI, Anti-HIST1H3G H3FH, Anti-HIST1H3H H3FK, Anti-HIST1H3I H3FF, Anti-HIST1H3J H3FJ, Anti-HIST2H3A HIST2H3C H3F2 H3FM HIST2H3D
About This Item
Recommended Products
biological source
mouse
conjugate
unconjugated
antibody form
purified immunoglobulin
antibody product type
primary antibodies
clone
5E5-G5, monoclonal
form
buffered aqueous solution
mol wt
antigen ~17 kDa
concentration
~1.0 mg/mL
technique(s)
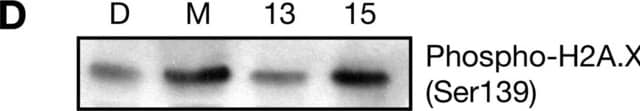
immunoblotting: 2-4 μg/mL using histones isolated from human HeLa cells.
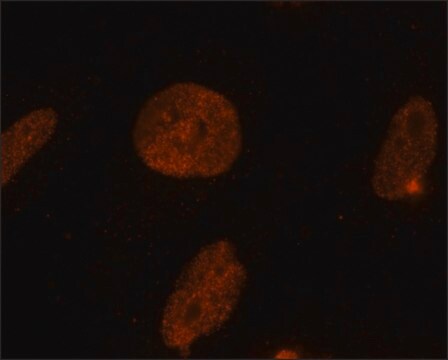
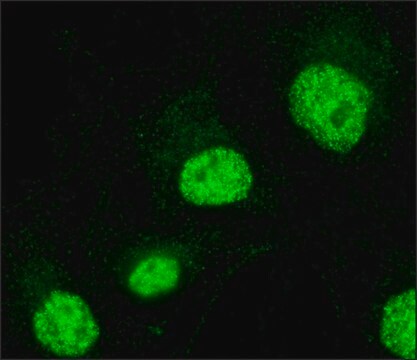
immunofluorescence: 1-2 μg/mL using human HeLa cells.
isotype
IgG1
shipped in
dry ice
storage temp.
−20°C
target post-translational modification
dimethylation (Lys9)
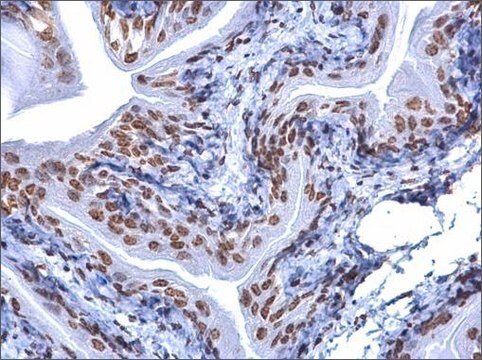
General description
Immunogen
Application
Biochem/physiol Actions
Physical form
Disclaimer
Not finding the right product?
Try our Product Selector Tool.
Storage Class Code
10 - Combustible liquids
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service