Target Identification with Hyas and Dione Kits
Introduction to Target Identification
Understanding how bioactive small molecules interact with healthy and diseased tissues is a critical step in the drug development. Identification of on- and off-targets can inform candidate selection and improve safety and efficacy of clinical programs. However, this key step is often a bottleneck in pharmaceutical development due to intrinsic challenges associated with target identification.
The current gold standard for small molecule target identification is a technique named photoaffinity labeling (PAL). In this method, the ligand under investigation is tethered to an alkyl diazirine, which decomposes under UV light irradiation to form a highly reactive carbene. This carbene can then insert into C-H and X-H bonds of proteins forming a stoichiometric cross-link allowing enrichment and identification.
Challenges in the Current Target Identification Method
Despite the widespread use of PAL, significant challenges remain that can make target identification a time consuming and laborious task for practitioners. In many cases, when the orientation of the alkyl diazirine is not optimal, >99% of the in situ generated carbene reacts only with water, leading to minimal crosslinking and significantly complicating analysis. One confounding aspect of this mechanism is that the hydrated product remains in the active site of the protein, precluding the entry of other diazirine bearing molecules (Figure 1). This property can lead to high background labeling and misidentification of targets. These issues are exacerbated when dealing with low copy number proteins or membrane bound proteins that are traditionally challenging to detect.
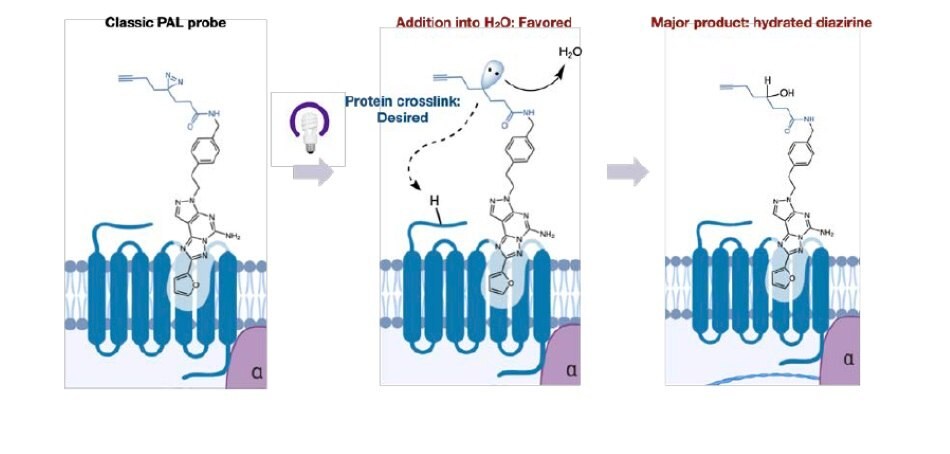
Figure 1.Classic PAL probes react with water leading to low crosslinking yields.
Hyas/Dione Micromapping (μMap) Kits for Photocatalytic Target Identification
Recently, Prof. MacMillan’s group at Princeton developed a method to address these issues associated with PAL based method. They questioned whether it would be possible to perform target ID in a catalytic manner, effectively increasing the amount of cross-linked protein, thereby amplifying the signal, and enabling the discovery of challenging protein targets. This was achieved by decoupling the cross-linker (diazirine) from the small molecule ligand, instead conjugating the ligand to an Ir-catalyst. This photosensitizer enables catalytic cross-linking by activating aryl diazirines (to form carbenes) following blue light irradiation. This photosensitization process is catalytic and allows temporal control of labeling and leads to a higher concentration of labeled peptides. By tethering these iridium catalysts to the small molecule under investigation, this method provides an extremely effective method for small molecule target identification for cases where conventional PAL probes are unsuccessful (Figure 2A).

Figure 2.(A) Catalytic diazirine activation leads to higher cross-linking signal for target identification. (B) Dione and Hyas catalysts for photocatalytic target identification.
We have partnered with the MacMillan lab to develop the Dione and Hyas kits, that allows users to rapidly conjugate iridium catalysts onto any small molecule of their choice in an operationally simple procedure (Figure 2B). The kits contain enough of the biotinylated diazirine warhead to perform 10 μMap experiments.
Key Advantages
- Facile incorporation of small molecules as an amide linkage, either with an amine or acid group
- Short labeling radius
- Benign method of activation (visible light)
- Catalytic signal amplification
References
To continue reading please sign in or create an account.
Don't Have An Account?