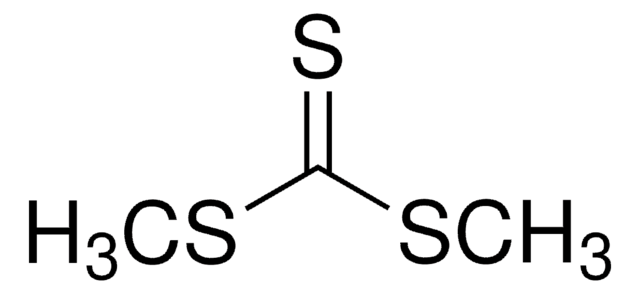
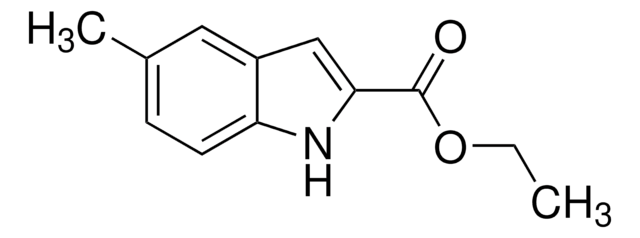
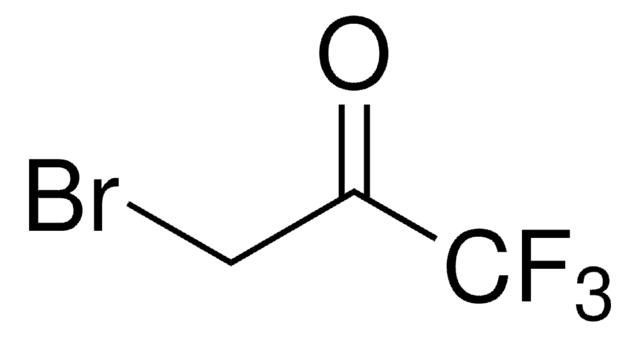
804347
3-nitro-N-[(3S)-tetrahydro-2-oxo-furanyl]-Benzeneacetamide
Synonym(s):
LasR inhibitor, LuxR activator
About This Item
Recommended Products
form
solid
storage temp.
−20°C
Application
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Related Content
Our laboratory pursues research at the chemistry-microbiology interface. We are deeply interested in the mechanisms by which bacteria sense each other, their environment, and the eukaryotic hosts on which and in which they may reside. One prominent pathway that we study is called quorum sensing, which allows bacteria to assess their local population density and initiate group behaviors at high cell (or “quorate”) density. This pathway allows, for example, many pathogens to amass in large populations prior to attacking their hosts. Bacteria use chemical signals for quorum sensing, and it is the concentration of these signals in a given environment that alerts the bacteria to their current cell number. We are interested in the structures of these signals and how we can reengineer them to either ablate or amplify quorum-sensing networks. Through synthesis and systematic screening, we have identified critical structural features of these signals and non-native functionality that we can install into the signals to tune their function. Thereby, we have developed non-native molecules that strongly inhibit or activate quorum-sensing pathways and modify infection processes. These compounds represent useful tools to explore the role of quorum sensing in many biological processes. We are applying them to both study fundamental aspects of quorum sensing pathways, and examine different types of infections in animals and plants.
Our laboratory pursues research at the chemistry-microbiology interface. We are deeply interested in the mechanisms by which bacteria sense each other, their environment, and the eukaryotic hosts on which and in which they may reside. One prominent pathway that we study is called quorum sensing, which allows bacteria to assess their local population density and initiate group behaviors at high cell (or “quorate”) density. This pathway allows, for example, many pathogens to amass in large populations prior to attacking their hosts. Bacteria use chemical signals for quorum sensing, and it is the concentration of these signals in a given environment that alerts the bacteria to their current cell number. We are interested in the structures of these signals and how we can reengineer them to either ablate or amplify quorum-sensing networks. Through synthesis and systematic screening, we have identified critical structural features of these signals and non-native functionality that we can install into the signals to tune their function. Thereby, we have developed non-native molecules that strongly inhibit or activate quorum-sensing pathways and modify infection processes. These compounds represent useful tools to explore the role of quorum sensing in many biological processes. We are applying them to both study fundamental aspects of quorum sensing pathways, and examine different types of infections in animals and plants.
Our laboratory pursues research at the chemistry-microbiology interface. We are deeply interested in the mechanisms by which bacteria sense each other, their environment, and the eukaryotic hosts on which and in which they may reside. One prominent pathway that we study is called quorum sensing, which allows bacteria to assess their local population density and initiate group behaviors at high cell (or “quorate”) density. This pathway allows, for example, many pathogens to amass in large populations prior to attacking their hosts. Bacteria use chemical signals for quorum sensing, and it is the concentration of these signals in a given environment that alerts the bacteria to their current cell number. We are interested in the structures of these signals and how we can reengineer them to either ablate or amplify quorum-sensing networks. Through synthesis and systematic screening, we have identified critical structural features of these signals and non-native functionality that we can install into the signals to tune their function. Thereby, we have developed non-native molecules that strongly inhibit or activate quorum-sensing pathways and modify infection processes. These compounds represent useful tools to explore the role of quorum sensing in many biological processes. We are applying them to both study fundamental aspects of quorum sensing pathways, and examine different types of infections in animals and plants.
Our laboratory pursues research at the chemistry-microbiology interface. We are deeply interested in the mechanisms by which bacteria sense each other, their environment, and the eukaryotic hosts on which and in which they may reside. One prominent pathway that we study is called quorum sensing, which allows bacteria to assess their local population density and initiate group behaviors at high cell (or “quorate”) density. This pathway allows, for example, many pathogens to amass in large populations prior to attacking their hosts. Bacteria use chemical signals for quorum sensing, and it is the concentration of these signals in a given environment that alerts the bacteria to their current cell number. We are interested in the structures of these signals and how we can reengineer them to either ablate or amplify quorum-sensing networks. Through synthesis and systematic screening, we have identified critical structural features of these signals and non-native functionality that we can install into the signals to tune their function. Thereby, we have developed non-native molecules that strongly inhibit or activate quorum-sensing pathways and modify infection processes. These compounds represent useful tools to explore the role of quorum sensing in many biological processes. We are applying them to both study fundamental aspects of quorum sensing pathways, and examine different types of infections in animals and plants.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service