CRISPR Cas9 Ribonucleoprotein (RNP) Application Note
CRISPR RNP-Based Genome Editing Applications
Although the CRISPR system can be delivered to cells via plasmids, direct introduction of Cas9 RNP strengthens and expands the applications of CRISPR genome modification technology by eliminating the possibility of plasmid DNA integration into the host genome. RNP complexes offer faster gene editing because the functional nuclease is immediately available in the cell and the complexes are quickly degraded and cleared making the editing activity short-lived. The reduced time of available RNP in the cells leads to reduced off-target effects.
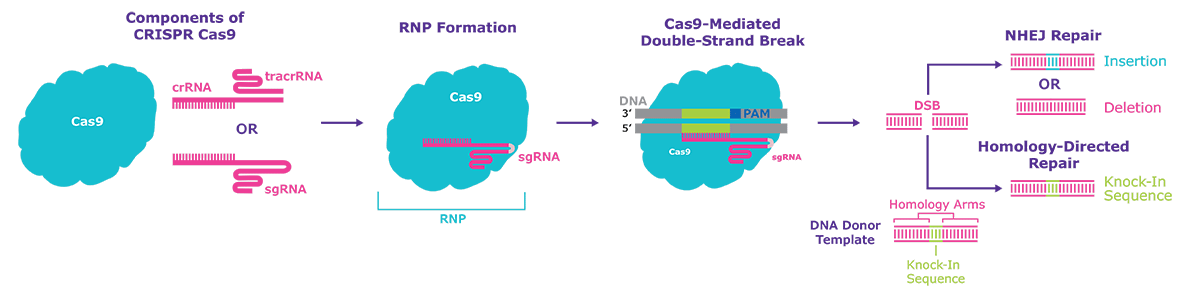
Figure 1.Schematic diagram of the CRISPR one-part and two-part systems with Cas9 protein, demonstrating the composition of the Ribonucleoprotein (RNP). After a double-strand break is created two possible repair pathways are taken by the cellular DNA machinery; Non-Homologous End Joining (NHEJ), Indel formation or Homology-Directed Repair (HDR) for targeted insertions or knock-ins.
Detailed Protocols
SygRNA® synthetic guides are compatible with a wide range of gene editing applications. Whether you need general use guidelines, nucleofection & microinjection of embryos, T cell editing, or iPSC protocols. You can also find help with downstream cleavage efficiency assessment, clone isolation & screening, and troubleshooting.
Primary T Cell Editing Using RNP
Ribonucleoprotein (RNP)-based genome editing experiments have never been easier than with SygRNA® synthetic guide RNAs and Sigma-Aldrich® Cas9 proteins, however some cells can be harder to edit than others. We have extensively validated our reagents and protocols to work in the most challenging of applications, including in primary T cells which present a number of significant challenges that need to be overcome when attempting to knock-in or knockout a construct or gene. However, with SygRNA® sgRNAs, editing in primary human T cells at efficiencies and specificities that exceed two-part guide systems can be achieved (Figure 2). In both primary T cells and PBMCs, SygRNA® sgRNAs can be introduced as highly specific paired nickases and Cas9 nickase, or as single site SpCas9 protein to get complete knockouts of the PD-1 gene, as measured by PD-1 expression and NGS (Figure 2). sgRNAs can be rapidly designed for any gene as either a single target RNA or as part of the paired nickase system, ideal for a variety of human cell types, including primary T-cells. Cas nucleases when delivered in protein format, pre-complexed with a single, modified guide RNA (gRNA), rather than encoded on a plasmid, allow researchers to bypass the historically limiting factors of using CRISPR in the therapeutic realm of off-targeting effects, low efficiency, and undesired cellular responses to foreign DNA. When you need to be certain, SygRNA® sgRNAs are the only guide RNAs that are fully backed by a 100% performance guarantee – predesigned and custom sequences.
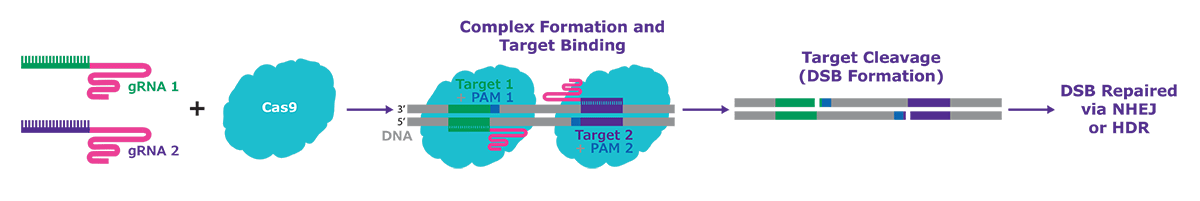
Figure 2.Editing with RNP Nickase systems, with only a single active cutting domain, and then delivering with proximal guide RNAs in a pair allows precise double stranded breaks, and provides hyper-specific and efficient genome editing in cell types that are more difficult to edit.
Proven High-performance Gene Editing in Mouse Embryos
Creating mouse models is a labor-intensive and expensive process, but it is critical for basic research in understanding health and disease. SygRNA® single guide RNA performance was tested at the highest level for genome engineering in mouse embryo applications by comparing our sgRNAs to other top providers at multiple genomic sites in mouse embryos.
Multiple replicates at each location in side by side comparisons revealed differences in editing efficiencies between identical sgRNA targets between Sigma-Aldrich® sgRNA and other top providers. Experiments conducted in parallel by an independent, prominent, mouse core facility show the superior performance of SygRNA® single guides in cutting efficiency. Additionally, there was no increase in toxicity to the embryo’s survivability. Collectively, the data shows that our highly active and pure sgRNAs provide superior gene editing in experiments conducted by an independent facility in comparison to competitors independent of microinjection or electroporation of the embryos.
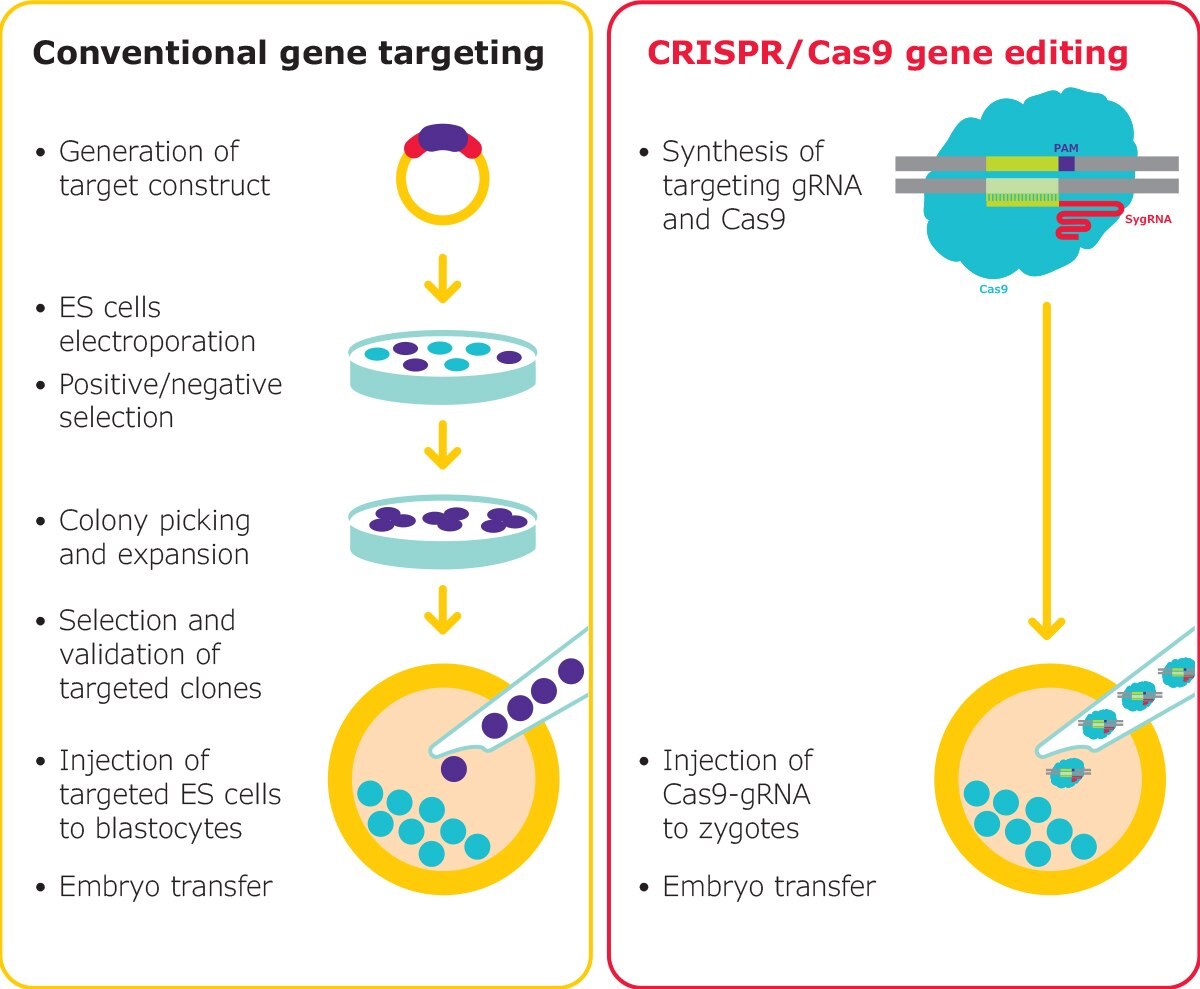
Figure 3.Gene editing in mouse embryos is traditionally a long process, however, RNP-based editing with CRISPR reduces the complexity. Mouse models can be generated rapidly, for nearly any target. SygRNA® is suitable for both microinjection and electroporation of embryos.
Related Products | |
|---|---|
| Product No. | Description |
| CAS9GFPPRO | Cas9-GFP Protein from Streptococcus pyogenes, fused with enhanced GFP, recombinant, expressed in E. coli, 3X NLS |
| ECAS9GFPPR | eSpCas9-GFP Protein from Streptococcus pyogenes with mutations conferring enhanced specificity, fused with enhanced GFP, recombinant, expressed in E. coli, 3X NLS |
| CAS9PROT | Cas9 Protein from Streptococcus pyogenes, recombinant, expressed in E. coli, 1X NLS |
| ESPCAS9PRO | eSpCas9 Protein from Streptococcus pyogenes with mutations conferring enhanced specificity, recombinant, expressed in E. coli, 1X NLS |
| DCAS9PROT | dCas9-3XFLAG™-Biotin Protein from Streptococcus pyogenes with D10A and H840A mutations, recombinant, expressed in E. coli, 1X NLS |
| CAS9D10APR | Cas9-D10A Nickase Protein from Streptococcus pyogenes with D10A mutation, recombinant, expressed in E. coli, 1X NLS |
| FNCAS9PROT | FnCas9 Protein from Francisella novicida, recombinant, expressed in E. coli, 1X NL |
| Product No. | Description |
|---|---|
| Custom Guide RNA | Order custom guide RNAs on line in various formats |
| TRACRRNA05N | SygRNA Cas9 Synthetic tracrRNA |
| TRACRRNAMOD | SygRNA Cas9 Synthetic modified tracrRNA |
| T1706 | TransIT®-CRISPR |
| XTG360-RO | X-tremeGENETM 360 Transfection Reagent |
| FNCAS9TRACR-5NMOL | SygRNA® FnCas9 tracrRNA |
| Embryo Culture Media | Order various types of mouse embryo culture media and mouse embryo tested reagents |
| Cell Culture | A complete catalog of cells and cell culture reagents |
Troubleshooting
If no cutting is observed and there is reason to suspect an experimental flaw is at fault, the following considerations may aid in troubleshooting the experiment.
| Suspected Issue | Solution |
|---|---|
| The Cas9 protein has denatured after long-term storage in dilution buffer. | The provided dilution buffer is only recommended for immediate use. For long-term storage, keep the protein lyophilized or resuspended in the provided Reconstitution solution at -20 ℃. |
| The Cas9 protein has been thawed and refrozen too many times. | The Cas9 protein has been shown to withstand several rounds of freezing and thawing without sacrificing cutting activity, but aliquoting the protein into smaller quantities upon resuspension will allow this potential issue to be avoided. |
| The crRNAs and tracrRNAs need to be annealed before complexing with the Cas9 protein. | Although an annealing step is generally not needed, it has shown to increase cutting in rare cases. To anneal the crRNA and tracrRNA, mix them in the desired ratio and incubate the mixture for 5 minutes at 95 ℃, then place the mixture on ice for 20 minutes. |
| The crRNAs and tracrRNAs are degraded. | Under normal cell culture conditions, synthetic RNA modifications are not needed for stabilization; however, for certain cell lines, this may be necessary. Modifications are available through us. |
| The transfection or nucleofection is not working or is too toxic. | For any transfection reagent or nucleofection, optimize the protocol for each cell line used. Refer to the manufacturer’s protocol for further assistance. |
| In vitro transcribed RNA is low quality or degraded. | For optimal performance, use only quality-verified IVT RNA. |
Network error: Failed to fetch
To continue reading please sign in or create an account.
Don't Have An Account?