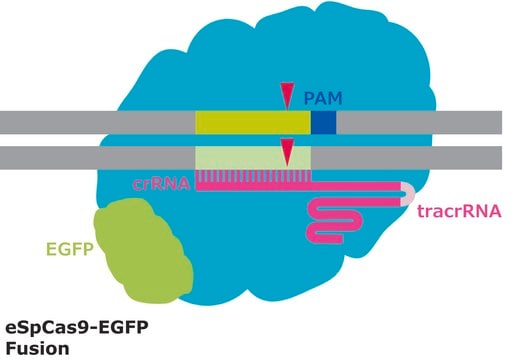
This system may only be used to inactivate genes. This item has an active Cas9 that cuts DNA, causing a deletion or an insertion after the cell repairs the break. This insertion or deletion will result in a frame shift, thereby stopping the correct translation of protein. This is a gene knockout. If a donor DNA template is added, homologous recombination occurs and the donor template is inserted into the DNA. CRISPR activation generally involves a dCas9 (inactive, dead Cas9) that has another functional protein domain fused to it that stimulates RNA transcription. The dCas9 fusion protein for gene activation is generally targeted to the transcription start site (TSS) of genes.
Recommended Products
Related Categories
General description
CRISPR Plant Cas9 products are intended for Agrobacterium-mediated plant transformation or biolistic microparticle bombardment or protoplast transformation. The products are based on the type IIA CRISPR-Cas9 derived from Streptococcus pyogenes. The native Cas9 coding sequence is codon optimized for expression in monocots and dicots, respectively. The monocot Cas9 constructs contain a monocot U6 promoter for sgRNA expression, and the dicot Cas9 constructs contain a dicot U6 promoter.
The plant selection markers include:
- hygromycin B resistance gene
- neomycin phosphotransferase gene
- bar gene (phosphinothricin acetyl transferase)
Application
- Inactivation of genes
- Target validation
- Site specific integration of gene of interest
- Gene replacement via HR
Features and Benefits
- Main advantages of CRISPR/Cas9 are in terms of simplicity, accessibility, cost and versatility.
- CRISPR/Cas9 system does not require any protein engineering steps, making it much more straightforward to test multiple gRNAs for each target gene
- Only 20 nt in the gRNA sequence need to be changed to confer a different target specificity
- Another advantage of CRISPR/Cas9 compared to ZFNs and TALENs is the ease of multiplexing. The simultaneous introduction of DSBs at multiple sites can be used to edit several genes at the same time. It can be particularly useful to knock out redundant genes or parallel pathways. Multiplex editing with the CRISPR/Cas9 system simply requires the monomeric Cas9 protein and any number of different sequence-specific gRNAs.
- CRISPR/Cas9 system can cleave methylated DNA in human cells allowing genomic modifications that are beyond the reach of the other nucleases. This has not been specifically explored in plants, it is reasonable to assume that the ability to cleave methylated DNA is intrinsic to the CRISPR/Cas9 system and not dependent on the target genome
Components
Keep reagent tubes closed when not in use.
Practice aseptic lab technique to avoid DNase contamination.
Principle
Other Notes
Legal Information
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
It looks like we've run into a problem, but you can still download Certificates of Analysis from our Documents section.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
-
Can this be used to activate genes rather than inactivation? If so, how can it be used to activate certain genes.
1 answer-
Helpful?
-
-
which strain of Agrobacterium tumefaciens is recommended when using p63, p55, p53 or p54 vector
1 answer-
Please navigate to the link https://www.sigmaaldrich.com/techservice, click on "Product Technical Inquiries" under the Products Section with all the required information so that a member of the Technical Service team can reach out to assist further. Thank you.
Helpful?
-
-
How can I use the CRISPR plant vectors for homologous recombination (HR)?
1 answer-
First, you need to design an HR DNA donor with the target of interest and the homologous arms. For Agrobacterium-mediated plant transformation, the HR DNA donor can be inserted into an appropriate restriction site, such as the EcoRI site or the HindIII site, within the T-DNA borders. For biolistics or protoplast plant transformation, the HR DNA donor can be inserted into XbaI, EcoRI, HindIII, or XhoI site. Alternatively the HR DNA donor can be co-delivered in a separate donor vector.
Helpful?
-
-
What is the promoter used for sgRNA (single guide RNA) expression?
1 answer-
In the monocot CRISPR vectors, it is a wheat U6 promoter. In the dicot CRISPR vectors, it is an Arabidopsis thaliana U6 promoter.
Helpful?
-
-
Can I use the monocot CRISPR plant vectors on dicots plants and the dicot CRISPR plant vectors on monocot plants?
1 answer-
Yes. But it is not recommended. The monocot CRISPR plant vectors are more suited for monocot plants, and the dicots CRISPR plant vectors are more suited for dicot plants.
Helpful?
-
-
What is the promoter used for the expression of the selection markers?
1 answer-
The 2X35S promoter is used in this case.
Helpful?
-
-
What antibiotics should I use for selection of the CRISPR Plant vectors in E. coli or Agrobacteria tumefaciens?
1 answer-
Kanamycin.
Helpful?
-
-
How is the Cas9 codon sequence optimized?
1 answer-
In the monocot CRISPR Plant vectors, the Cas9 codon optimization is based on Zea mays. In the dicot CRISPR Plant vectors, the Cas9 codon optimization is based on Arabidopsis thaliana.
Helpful?
-
-
Can one use an Agrobacterium-mediated transformation CRISPR vector for biolistics or protoplast plant transformation?
1 answer-
Yes. However, the biolistics/protoplast CRISPR vectors cannot be used for Agrobacterium-mediated plant transformation.
Helpful?
-
-
1. What type of Cas9 is used in the CRISPR Plant vectors?
1 answer-
Type IIA Cas9 derived from Streptococcus pyogenes.
Helpful?
-
Active Filters
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service