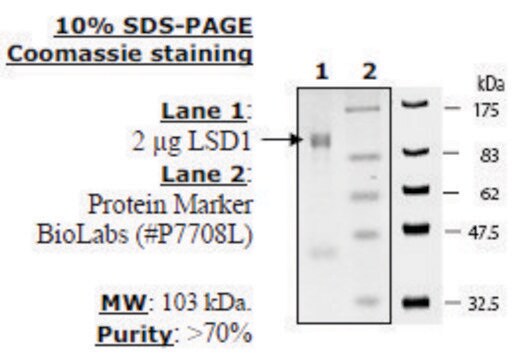
SRP0125
LSD1 substrate (Di-methylated K4_H3)
≥90% (HPLC), aqueous solution
Synonym(s):
HIST1H3E, Histone H3.1
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
12352204
NACRES:
NA.32
Recommended Products
product name
LSD1 substrate (Di-methylated K4_H3), ≥90% (HPLC)
biological source
human
Assay
≥90% (HPLC)
form
aqueous solution
mol wt
2283 Da
packaging
pkg of 500UL
concentration
0.55 mg/mL
UniProt accession no.
shipped in
dry ice
storage temp.
−70°C
Gene Information
human ... HIST1H3E(8353)
General description
Lysine-specific demethylase 1 (LSD1) acts on mono- and dimethylated histones and demethylates them. It has a role in embryogenesis, cell proliferation, spermatogenesis, adipogenesis and chromosomal segregation. Histone H3 peptide methylated at Lysine 4 is a substrate for LSD1. Histone cluster 1 H3 family member E (HIST1H3E) is a variant of histone 3 and is also called as the replicative histone. It is mainly expressed in the S-phase of the cell cycle. The gene encoding this protein is localized on human chromosome 6p22.
Application
Study enzyme kinetics and screen small molecular inhibitors of LSD1 for drug discovery and HTS applications
Biochem/physiol Actions
Histones have an important role in the organization and modification of chromatin. Histone cluster 1 H3 family member E (HIST1H3E) has a role in DNA synthesis during DNA replication and might also be involved in DNA repair. It is deposited at the chromatin at the time of DNA replication-associated chromatin assembly. During herpes simplex virus 1 (HSV-1) infection, differential movement of HIST1H3E is associated with the assembly of viral chromatin.
Physical form
Supplied as 500 uL of water solution at 200 uM concetration.
Preparation Note
Thaw on ice. Upon first thaw, briefly spin tube to recover full content of the tube. Aliquot into single use aliquots. Store remaining product in aliquots at -70°C. Note: Avoid freeze/thaw cycles.
Storage Class Code
10 - Combustible liquids
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Yingwei Chen et al.
Critical reviews in eukaryotic gene expression, 22(1), 53-59 (2012-02-22)
Lysine-specific demethylase 1 (LSD1), the first identified histone demethylase, was belonged to the superfamily of the flavin adenine dinucleotide (FAD)-dependent amine oxidases. LSD1 specifically demethylates mono- or dimethylated dimethylated histone H3 lysine4 (H3K4) and H3 lysine 9 (H3K9) via a
Kristen L Conn et al.
PLoS pathogens, 9(10), e1003695-e1003695 (2013-10-17)
During lytic infections, HSV-1 genomes are assembled into unstable nucleosomes. The histones required for HSV-1 chromatin assembly, however, are in the cellular chromatin. We have shown that linker (H1) and core (H2B and H4) histones are mobilized during HSV-1 infection
Kami Ahmad et al.
Molecular cell, 9(6), 1191-1200 (2002-06-28)
Two very similar H3 histones-differing at only four amino acid positions-are produced in Drosophila cells. Here we describe a mechanism of chromatin regulation whereby the variant H3.3 is deposited at particular loci, including active rDNA arrays. While the major H3
Eric I Campos et al.
Nature structural & molecular biology, 17(11), 1343-1351 (2010-10-19)
The mechanism by which newly synthesized histones are imported into the nucleus and deposited onto replicating chromatin alongside segregating nucleosomal counterparts is poorly understood, yet this program is expected to bear on the putative epigenetic nature of histone post-translational modifications.
Alejandra Loyola et al.
Molecular cell, 24(2), 309-316 (2006-10-21)
Histone posttranslational modifications (PTMs) and sequence variants regulate genome function. Although accumulating evidence links particular PTM patterns with specific genomic loci, our knowledge concerning where and when these PTMs are imposed remains limited. Here, we find that lysine methylation is
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service