推荐产品
材料
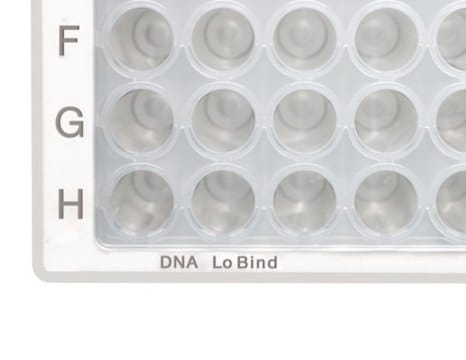
clear wells
colorless wells
conical bottom
polypropylene
white plate
無菌
non-sterile
特點
lid: no
包裝
pkg of 120 ea (10 bags × 12 plates)
製造商/商標名
Eppendorf® 951032808
容量
1000 μL
孔容積
1000 μL
顏色
white border
適合性
suitable for PCR
suitable for molecular biology
正在寻找类似产品? 访问 产品对比指南
相关类别
一般說明
Deepwell Plate 96/1000µL, DNA LoBind, wells clear, 1,000 µL, LoBind, PCR clean, white, 20 plates (5 bags × 4 plates)
- Eppendorf LoBind material ensures optimized sample recovery for improved assay results
- Free of surface coating (e.g. silicone) to minimize the risk of sample interference
- Lot-certified PCR clean purity grade: free of human DNA, DNase, RNase and PCR inhibitors
- Available in tube, microplate, and deepwell plate formats for easy-up scaling
- High-contrast Unique OptiTrack® matrix: up to 30 % faster sample identification and fewer pipetting errors
- RecoverMax well design: optimized well geometry for minimal remaining/dead volume and excellent mixing properties
- Raised well rims and a smooth surface ensure reliable sealing
特點和優勢
- Eppendorf LoBind material ensures optimized sample recovery for improved assay results
- Free of surface coating (e.g. silicone) to minimize the risk of sample interference
- Lot-certified PCR clean purity grade: free of human DNA, DNase, RNase and PCR inhibitors
- Available in tube, microplate, and deepwell plate formats for easy-up scaling
- High-contrast Unique OptiTrack® matrix: up to 30 % faster sample identification and fewer pipetting errors
- RecoverMax® well design: optimized well geometry for minimal remaining/dead volume and excellent mixing properties
- Raised well rims and a smooth surface ensure reliable sealing
法律資訊
Eppendorf is a registered trademark of Eppendorf AG
OptiTrack is a registered trademark of Eppendorf AG
RecoverMax is a registered trademark of Eppendorf AG
Kelly Hodge et al.
Journal of proteomics, 88, 92-103 (2013-03-19)
Mass spectrometry, in the past five years, has increased in speed, accuracy and use. With the ability of the mass spectrometers to identify increasing numbers of proteins the identification of undesirable peptides (those not from the protein sample) has also
Arzu Umar et al.
Proteomics, 7(2), 323-329 (2006-12-14)
Proteomics assays hold great promise for unraveling molecular events that underlie human diseases. Effective analysis of clinical samples is essential, but this task is considerably complicated by tissue heterogeneity. Laser capture microdissection (LCM) can be used to selectively isolate target
Byung-Gyu Kim et al.
Proteomics, 6(4), 1166-1174 (2006-01-20)
Runx2 is a key transcription factor in osteoblast differentiation, and its activity is regulated by fibroblast growth factors (FGFs). Craniosynostosis, characterized by premature suture closure, results from mutations that generate constitutively active FGF receptors (FGFRs). We previously showed that FGF/FGFR-activated
Cláudia P Grou et al.
The Journal of biological chemistry, 283(21), 14190-14197 (2008-03-25)
According to current models of peroxisomal biogenesis, newly synthesized peroxisomal matrix proteins are transported into the organelle by Pex5p. Pex5p recognizes these proteins in the cytosol, mediates their membrane translocation, and is exported back into the cytosol in an ATP-dependent
Graziano Colombo et al.
Free radical biology & medicine, 52(9), 1584-1596 (2012-03-06)
Cigarette smoke, a complex mixture of over 7000 chemicals, contains many components capable of eliciting oxidative stress, which may induce smoking-related disorders, including oral cavity diseases. In this study, we investigated the effects of whole (mainstream) cigarette smoke on human
我们的科学家团队拥有各种研究领域经验,包括生命科学、材料科学、化学合成、色谱、分析及许多其他领域.
联系技术服务部门