E6412
Cellobiohydrolase I from Hypocrea jecorina
0.13 U/mg, recombinant, expressed in corn
Synonym(s):
Cel7A, Cellobiosidase, Cellulase
Sign Into View Organizational & Contract Pricing
All Photos(2)
About This Item
Recommended Products
recombinant
expressed in corn
Quality Level
form
liquid
specific activity
0.13 U/mg
greener alternative product characteristics
Design for Energy Efficiency
Learn more about the Principles of Green Chemistry.
sustainability
Greener Alternative Product
greener alternative category
shipped in
dry ice
storage temp.
−20°C
General description
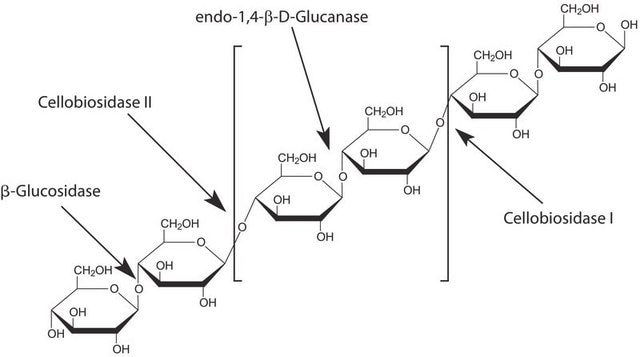
Cellubiohydrolase I is an enzyme present in many fungi, but particularly wood rot fungi. It is a monomer of 53 kDa with a catalytic domain and a cellulose binding domain. The reaction adds water to the glucose bonds in cellulose (non-reducing ends of the chain), yielding cellobiose.
We are committed to bringing you Greener Alternative Products, which adhere to one or more of The 12 Principles of Greener Chemistry. This product has been enhanced for energy efficiency. Find details here.
Application
Cellobiohydrolase I can be used in combination with endocellulases and b-glucosidase to produce glucose from cellulose.
Biochem/physiol Actions
Cellobiohydrolase (CBH) is a cellulase which degrades cellulose by hydrolysing the 1,4-β-D-glycosidic bonds. CBH is an exocellulase which cleaves two to four units from the ends of cellulose. CBH I cleaves progressively from the reducing end. CBH I is commonly used in detergents for cleaning textiles. Its ezymatic activity ranges from 37° C to 50° C, with its optimal temperature being approximately 45° C. The optimum pH for the enzyme is 5-6.
Unit Definition
Unit Definition: A unit will turn over 1 nmole of methyl-umbelliferyl beta-D cellobioside per min at pH 5 at 50° C.
Physical form
Provided as an ammonium sulfate precipitate with the source as recombinant maize.
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Resp. Sens. 1
Storage Class Code
10 - Combustible liquids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Lot/Batch Number
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Larissa C Textor et al.
The FEBS journal, 280(1), 56-69 (2012-11-02)
Aiming to contribute toward the characterization of new, biotechnologically relevant cellulolytic enzymes, we report here the first crystal structure of the catalytic core domain of Cel7A (cellobiohydrolase I) from the filamentous fungus Trichoderma harzianum IOC 3844. Our structural studies and
Svein J Horn et al.
Methods in enzymology, 510, 69-95 (2012-05-23)
Natural cellulolytic enzyme systems as well as leading commercial cellulase cocktails are dominated by enzymes that degrade cellulose chains in a processive manner. Despite the abundance of processivity among natural cellulases, the molecular basis as well as the biotechnological implications
Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose.
Christina M Payne et al.
Proceedings of the National Academy of Sciences of the United States of America, 110(36), 14646-14651 (2013-08-21)
Plant cell-wall polysaccharides represent a vast source of food in nature. To depolymerize polysaccharides to soluble sugars, many organisms use multifunctional enzyme mixtures consisting of glycoside hydrolases, lytic polysaccharide mono-oxygenases, polysaccharide lyases, and carbohydrate esterases, as well as accessory, redox-active
Daguan Nong et al.
Biomedical optics express, 12(6), 3253-3264 (2021-07-06)
We describe a multimodal microscope for visualizing processive enzymes moving on immobilized substrates. The instrument combines interference reflection microscopy (IRM) with multi-wavelength total internal reflectance fluorescence microscopy (TIRFM). The microscope can localize quantum dots with a precision of 2.8 nm at
Naohisa Sugimoto et al.
Langmuir : the ACS journal of surfaces and colloids, 28(40), 14323-14329 (2012-09-07)
Cellobiohydrolases (CBHs) hydrolyzing crystalline cellulose share a two-domain structure of catalytic domain (CD) and cellulose-binding domain (CBD). To focus on the binding characteristics of CBD, we analyzed the adsorption of fusion protein of fungal family 1 CBD from Trichoderma reesei
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service