Wichtige Dokumente
930571
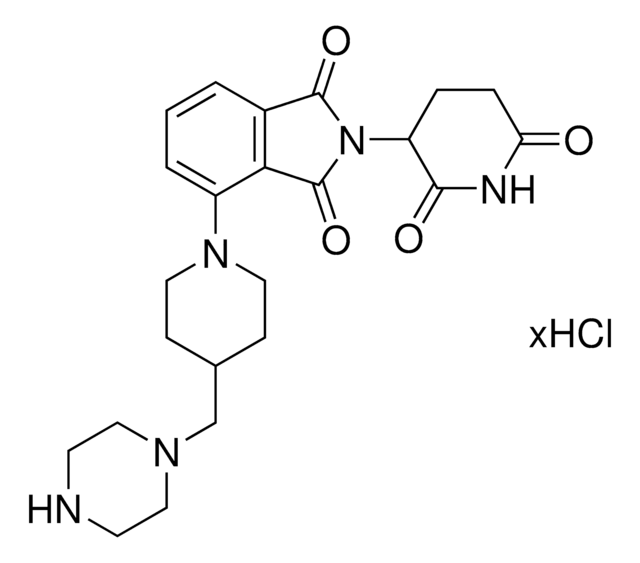
Pomalidomide-piperidine-carboxylic acid
≥95%
Synonym(e):
1-[2-(2,6-Dioxo-3-piperidinyl)-2,3-dihydro-1,3-dioxo-1H-isoindol-5-yl], 1-[2-(2,6-Dioxo-3-piperidyl)-1,3-dioxo-isoindolin-5-yl]piperidine-4-carboxylic acid, 4-Piperidinecarboxylic acid
About This Item
Empfohlene Produkte
ligand
pomalidomide
Qualitätsniveau
Assay
≥95%
Form
powder
Funktionelle Gruppe
carboxylic acid
Lagertemp.
2-8°C
SMILES String
O=C(O)C1CCN(C2=CC=C3C(=O)N(C(=O)C3=C2)C4C(=O)NC(=O)CC4)CC1
Verwandte Kategorien
Anwendung
Technology Spotlight: Degrader Building Blocks for Targeted Protein Degradation
Protein Degrader Building Blocks
Sonstige Hinweise
Destruction of DNA-Binding Proteins by Programmable Oligonucleotide PROTAC (O′PROTAC): Effective Targeting of LEF1 and ERG
Small-Molecule PROTACS: New Approaches to Protein Degradation
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Impact of linker length on the activity of PROTACs
Signalwort
Danger
H-Sätze
Gefahreneinstufungen
Repr. 1B
Lagerklassenschlüssel
6.1C - Combustible acute toxic Cat.3 / toxic compounds or compounds which causing chronic effects
WGK
WGK 3
Hier finden Sie alle aktuellen Versionen:
Analysenzertifikate (COA)
Die passende Version wird nicht angezeigt?
Wenn Sie eine bestimmte Version benötigen, können Sie anhand der Lot- oder Chargennummer nach einem spezifischen Zertifikat suchen.
Besitzen Sie dieses Produkt bereits?
In der Dokumentenbibliothek finden Sie die Dokumentation zu den Produkten, die Sie kürzlich erworben haben.
Unser Team von Wissenschaftlern verfügt über Erfahrung in allen Forschungsbereichen einschließlich Life Science, Materialwissenschaften, chemischer Synthese, Chromatographie, Analytik und vielen mehr..
Setzen Sie sich mit dem technischen Dienst in Verbindung.