927554
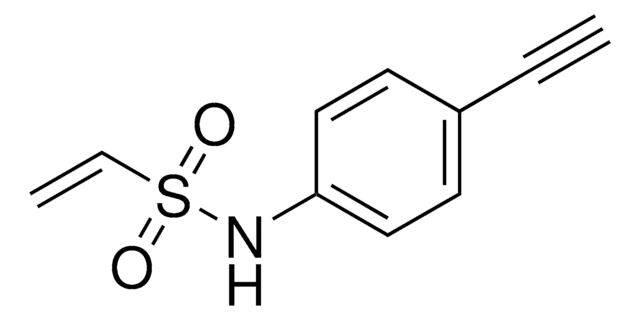
MI-alkyne
≥95%
Synonym(s):
N-(2-(2,5-Dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl)hex-5-ynamide
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
Empirical Formula (Hill Notation):
C12H14N2O3
Molecular Weight:
234.25
UNSPSC Code:
12352101
NACRES:
NA.21
Recommended Products
Application
MI-alkyne is a Michael acceptor probe that can be used to lable cysteines. A method was developed using cysteine-reactive compounds including this one to allow for unbiased analysis of proteomic data in quantitave applications (Zanon et al. 2021). The method uses light or heavy labeling with the isotopically labelled desthiobiotin azide (isoDTB) tag for mass spectrometry analysis (Zanon et al. 2020). Analysis then uses the isotopic tandem orthogonal proteolysis activity-based protein profiling (isoTOP-ABPP) workflow (Weerapana et al. 2010, Backus et al. 2016).
related product
Product No.
Description
Pricing
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Lot/Batch Number
Sorry, we don't have COAs for this product available online at this time.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Profiling the proteome-wide selectivity of diverse electrophiles.
Zanon P RA, et al.
ChemRxiv : the preprint server for chemistry (2021)
Patrick R A Zanon et al.
Angewandte Chemie (International ed. in English), 59(7), 2829-2836 (2019-11-30)
Rapid development of bacterial resistance has led to an urgent need to find new druggable targets for antibiotics. In this context, residue-specific chemoproteomic approaches enable proteome-wide identification of binding sites for covalent inhibitors. Described here are easily synthesized isotopically labeled
Eranthie Weerapana et al.
Nature, 468(7325), 790-795 (2010-11-19)
Cysteine is the most intrinsically nucleophilic amino acid in proteins, where its reactivity is tuned to perform diverse biochemical functions. The absence of a consensus sequence that defines functional cysteines in proteins has hindered their discovery and characterization. Here we
Keriann M Backus et al.
Nature, 534(7608), 570-574 (2016-06-17)
Small molecules are powerful tools for investigating protein function and can serve as leads for new therapeutics. Most human proteins, however, lack small-molecule ligands, and entire protein classes are considered 'undruggable'. Fragment-based ligand discovery can identify small-molecule probes for proteins
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service