764213
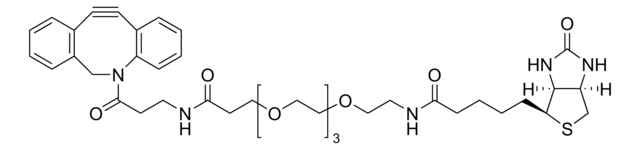
Biotin-PEG4-alkyne
for copper catalyzed click labeling
Synonym(s):
Polyethylene glycol, Acetylene-PEG4-biotin conjugate
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
Empirical Formula (Hill Notation):
C21H35N3O6S
CAS Number:
Molecular Weight:
457.58
UNSPSC Code:
12161502
PubChem Substance ID:
NACRES:
NA.22
Recommended Products
Quality Level
Assay
95%
form
solid
reaction suitability
reaction type: click chemistry
mp
55-64 °C
storage temp.
−20°C
SMILES string
O=C(NCCOCCOCCOCCOCC#C)CCCC[C@@H](SC1)[C@@]2([H])[C@]1([H])NC(N2)=O
InChI
1S/C21H35N3O6S/c1-2-8-27-10-12-29-14-15-30-13-11-28-9-7-22-19(25)6-4-3-5-18-20-17(16-31-18)23-21(26)24-20/h1,17-18,20H,3-16H2,(H,22,25)(H2,23,24,26)/t17-,18-,20-/m1/s1
InChI key
SKMJWNZZFUDLKQ-QWFCFKBJSA-N
Related Categories
Application
Biotin-PEG4-alkyne may be used for the modification of 4-azidophenylalanine (AzPhe) silk fibroin via bioorthogonal azide–alkyne cycloaddition reaction for developing photopatternable protein material.
Biotinylation reagent for labeling azide containing molecules or biomolecules using copper-catalyzed 1,3 dipolar cycloaddition click chemistry. The alkyne group reacts with azides to form a stable triazole linkage, facilitating the introduction of biotin into your azide modified system of interest.
Automate your Biotin tagging with Synple Automated Synthesis Platform (SYNPLE-SC002)
Automate your Biotin tagging with Synple Automated Synthesis Platform (SYNPLE-SC002)
Other Notes
Chemoproteomic Profiling of Gut Microbiota-Associated Bile Salt Hydrolase Activity
Fluorescent Heterotelechelic Single-Chain Polymer Nanoparticles: Synthesis, Spectroscopy, and Cellular Imaging
Arginine-Selective Chemical Labeling Approach for Identification and Enrichment of Reactive Arginine Residues in Proteins
Selective Imaging of Gram-Negative and Gram-Positive Microbiotas in the Mouse Gut
Metabolic Oligosaccharide Engineering with Alkyne Sialic Acids Confers Neuraminidase Resistance and Inhibits Influenza Reproduction
A Modular Probe Strategy for Drug Localization, Target Identification and Target Occupancy Measurement on Single Cell Level
Fluorescent Heterotelechelic Single-Chain Polymer Nanoparticles: Synthesis, Spectroscopy, and Cellular Imaging
Arginine-Selective Chemical Labeling Approach for Identification and Enrichment of Reactive Arginine Residues in Proteins
Selective Imaging of Gram-Negative and Gram-Positive Microbiotas in the Mouse Gut
Metabolic Oligosaccharide Engineering with Alkyne Sialic Acids Confers Neuraminidase Resistance and Inhibits Influenza Reproduction
A Modular Probe Strategy for Drug Localization, Target Identification and Target Occupancy Measurement on Single Cell Level
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Masanobu Nagano et al.
Bioorganic & medicinal chemistry, 25(21), 5952-5961 (2017-10-11)
Vaccination is a reliable method of prophylaxis and a crucial measure for public health. However, the majority of vaccines cannot be administered orally due to their degradation in the harsh gut environment or inability to cross the GI tract. In
Maheshika S K Wanigasekara et al.
ACS omega, 3(10), 14229-14235 (2019-08-29)
Modification of arginine residues using dicarbonyl compounds is a common method to identify functional or reactive arginine residues in proteins. Arginine undergoes several kinds of posttranslational modifications in these functional residues. Identifying these reactive residues confidently in a protein or
Nrf1 can be processed and activated in a proteasome-independent manner.
Vangala JR, et al.
Current Biology, 26(18), R834-R835 (2016)
Lili Lu et al.
Angewandte Chemie (International ed. in English), 54(33), 9679-9682 (2015-06-24)
Glycosylphosphatidylinositol (GPI) anchoring of proteins to the cell surface is important for various biological processes, but GPI-anchored proteins are difficult to study. An effective strategy was developed for the metabolic engineering of cell-surface GPIs and GPI-anchored proteins by using inositol
Anna Rutkowska et al.
ACS chemical biology, 11(9), 2541-2550 (2016-07-08)
Late stage failures of candidate drug molecules are frequently caused by off-target effects or inefficient target engagement in vivo. In order to address these fundamental challenges in drug discovery, we developed a modular probe strategy based on bioorthogonal chemistry that
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service


![Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine 97%](/deepweb/assets/sigmaaldrich/product/structures/179/695/86a721c8-2a4c-4e4f-bc36-6276ce7a941f/640/86a721c8-2a4c-4e4f-bc36-6276ce7a941f.png)