추천 제품
생물학적 소스
rabbit
Quality Level
항체 형태
serum
항체 생산 유형
primary antibodies
클론
polyclonal
종 반응성
yeast, Saccharomyces cerevisiae, human
종 반응성(상동성에 의해 예측)
vertebrates (most common)
제조업체/상표
Upstate®
기술
ChIP: suitable (ChIP-seq)
dot blot: suitable
western blot: suitable
NCBI 수납 번호
UniProt 수납 번호
배송 상태
dry ice
타겟 번역 후 변형
acetylation (Lys18)
유전자 정보
human ... H3C1(8350)
일반 설명
Histone H3 is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N terminal tail H3 is involved with the structure of the nucleosomes of the ′beads on a string′ structure.
Acetylation of histone H3 occurs at several different lysine positions in the histone tail and is performed by a family of enzymes known as Histone Acetyl Transferases (HATs).
Acetylation of histone H3 occurs at several different lysine positions in the histone tail and is performed by a family of enzymes known as Histone Acetyl Transferases (HATs).
특이성
Recognizes Histone H3 acetylated on lysine 18.
면역원
KLH-conjugated, synthetic peptide (GKAPRAcKQLASK-C) corresponding to amino acids 13-23 of yeast Histone H3 acetylated on lysine 18with a C-terminal cysteine added for conjugation purposes
애플리케이션
Chromatin Immunoprecipitation:
Representative lot data.
Sonicated chromatin prepared from HeLa cells (1 X 10E6 cell equivalents per IP) were subjected to chromatin immunoprecipitation using 2 µL of either Normal Rabbit Serum , or 2 µL of Anti-Acetyl-Histone H3 (Lys18)and the Magna ChIP A Kit (Cat. # 17-610). Successful immunoprecipitation of Acetyl-Histone H3 (Lys18) associated DNA fragments was verified by qPCR using Control Primers specific for the human GAPDH promoter region as a positive locus, and β-globin primers as a negative locus. Data is presented as percent input of each IP sample relative to input chromatin for each amplicon and ChIP sample as indicated.
Please refer to the EZ-Magna ChIP A (Cat. # 17-408) or EZ-ChIP (Cat. # 17-371) protocol for experimental details.
ChIP-Sequencing:
Representative lot data. Chromatin immunoprecipitation was performed using the Magna ChIP HiSens kit (cat# 17-10460), 2 μg of anti-acetyl-Histone H3 (Lys18) antibody (cat# 07-354), 20 µL Protein A/G beads , and 1e6 crosslinked HeLa cell chromatin followed by DNA purification using magnetic beads. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on Illumina HiSeq instrument. An excess of ten million reads from FastQ files were mapped using Bowtie (http://bowtie-bio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files.
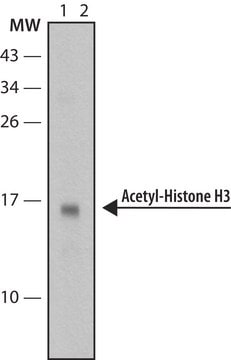
Western Blot Analysis:
Representative lot data.
Recombinant histone H3 (lane 1, Catalog # 14-494) and acid extracts from sodium butyrate treated (lane 2) and untreated (lane 3) HeLa cells (Catalog # 17-305) were probed with anti acetyl- Histone H3 (Lys18) (1:10,000 dilution).
Arrow indicates acetyl histone H3 (Lys18) (17 kDa).
Dot Blot:
Representative lot data.
40 ng and 4ng amounts of histone peptides with various modifications (see table 1) were transferred to PVDF membrane and probed with Anti-Acetyl-Histone H3 (Lys18) antibody (1:5000 dilution). Proteins were visualized using a goat anti-rabbit IgG conjugated to HRP and a chemiluminescence detection system. Image from a 60 second exposure is shown.
Representative lot data.
Sonicated chromatin prepared from HeLa cells (1 X 10E6 cell equivalents per IP) were subjected to chromatin immunoprecipitation using 2 µL of either Normal Rabbit Serum , or 2 µL of Anti-Acetyl-Histone H3 (Lys18)and the Magna ChIP A Kit (Cat. # 17-610). Successful immunoprecipitation of Acetyl-Histone H3 (Lys18) associated DNA fragments was verified by qPCR using Control Primers specific for the human GAPDH promoter region as a positive locus, and β-globin primers as a negative locus. Data is presented as percent input of each IP sample relative to input chromatin for each amplicon and ChIP sample as indicated.
Please refer to the EZ-Magna ChIP A (Cat. # 17-408) or EZ-ChIP (Cat. # 17-371) protocol for experimental details.
ChIP-Sequencing:
Representative lot data. Chromatin immunoprecipitation was performed using the Magna ChIP HiSens kit (cat# 17-10460), 2 μg of anti-acetyl-Histone H3 (Lys18) antibody (cat# 07-354), 20 µL Protein A/G beads , and 1e6 crosslinked HeLa cell chromatin followed by DNA purification using magnetic beads. Libraries were prepared from Input and ChIP DNA samples using standard protocols with Illumina barcoded adapters, and analyzed on Illumina HiSeq instrument. An excess of ten million reads from FastQ files were mapped using Bowtie (http://bowtie-bio.sourceforge.net/manual.shtml) following TagDust (http://genome.gsc.riken.jp/osc/english/dataresource/) tag removal. Peaks were identified using MACS (http://luelab.dfci.harvard.edu/MACS/), with peaks and reads visualized as a custom track in UCSC Genome Browser (http://genome.ucsc.edu) from BigWig and BED files.
Western Blot Analysis:
Representative lot data.
Recombinant histone H3 (lane 1, Catalog # 14-494) and acid extracts from sodium butyrate treated (lane 2) and untreated (lane 3) HeLa cells (Catalog # 17-305) were probed with anti acetyl- Histone H3 (Lys18) (1:10,000 dilution).
Arrow indicates acetyl histone H3 (Lys18) (17 kDa).
Dot Blot:
Representative lot data.
40 ng and 4ng amounts of histone peptides with various modifications (see table 1) were transferred to PVDF membrane and probed with Anti-Acetyl-Histone H3 (Lys18) antibody (1:5000 dilution). Proteins were visualized using a goat anti-rabbit IgG conjugated to HRP and a chemiluminescence detection system. Image from a 60 second exposure is shown.
Use Anti-acetyl-Histone H3 (Lys18) Antibody (Rabbit Polyclonal Antibody) validated in ChIP, WB to detect acetyl-Histone H3 (Lys18) also known as H3K18Ac, Histone H3 (acetyl K18).
품질
routinely evaluated by immunoblot on acid extracts from sodium butyrate treated HeLa cells
표적 설명
17 kDa
결합
Replaces: 04-1107
물리적 형태
100 μLof rabbit serum containing 0.05% sodium azide and 30% glycerol.
기타 정보
Concentration: Please refer to the Certificate of Analysis for the lot-specific concentration.
법적 정보
UPSTATE is a registered trademark of Merck KGaA, Darmstadt, Germany
Not finding the right product?
Try our 제품 선택기 도구.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point (°F)
Not applicable
Flash Point (°C)
Not applicable
시험 성적서(COA)
제품의 로트/배치 번호를 입력하여 시험 성적서(COA)을 검색하십시오. 로트 및 배치 번호는 제품 라벨에 있는 ‘로트’ 또는 ‘배치’라는 용어 뒤에서 찾을 수 있습니다.
Inducible activation of Cre recombinase in adult mice causes gastric epithelial atrophy, metaplasia and regenerative changes in the absence of "floxed" alleles.
Huh WJ, Mysorekar IU, Mills JC
American Journal of Physiology: Gastrointestinal and Liver Physiology null
Paola Y Bertucci et al.
Nucleic acids research, 41(12), 6072-6086 (2013-05-04)
Steroid receptors were classically described for regulating transcription by binding to target gene promoters. However, genome-wide studies reveal that steroid receptors-binding sites are mainly located at intragenic regions. To determine the role of these sites, we examined the effect of
Nucleosome competition reveals processive acetylation by the SAGA HAT module.
Ringel, AE; Cieniewicz, AM; Taverna, SD; Wolberger, C
Proceedings of the National Academy of Sciences of the USA null
Jason S Patzlaff et al.
Experimental cell research, 316(13), 2123-2135 (2010-05-11)
Histone acetylation is a key modification that regulates chromatin accessibility. Here we show that treatment with butyrate or other histone deacetylase (HDAC) inhibitors does not induce histone hyperacetylation in metaphase-arrested HeLa cells. When compared to similarly treated interphase cells, acetylation
H2B- and H3-specific histone deacetylases are required for DNA methylation in Neurospora crassa.
Smith, KM; Dobosy, JR; Reifsnyder, JE; Rountree, MR; Anderson, DC; Green, GR; Selker, EU
Genetics null
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.