MBD0051
Yeast FISH probe-Cy3
Probe for fluorescence in situ hybridization (FISH), 20µM in water
Synonym(s):
PF2-Cy3 probe
Sign Into View Organizational & Contract Pricing
All Photos(4)
About This Item
UNSPSC Code:
12352200
NACRES:
NA.54
Recommended Products
Quality Level
technique(s)
FISH: suitable
fluorescence
λex 550 nm; λem 570 nm (Cy3)
shipped in
dry ice
storage temp.
−20°C
General description
FISH (Fluorescent in Situ Hybridization) is based on the hybridization of fluorescently labeled oligonucleotide probes to a specific complementary DNA or RNA sequence in whole and intact cells. Microbial FISH allows the visualization, identification, and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples.
FISH can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (e.g., blood and other tissues), microbial ecology (biofilms, aquatic systems) and plants. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control probes (MBD0034/35) that accompany the specific probe of interest.
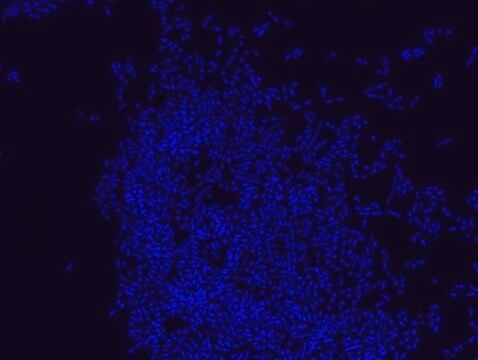
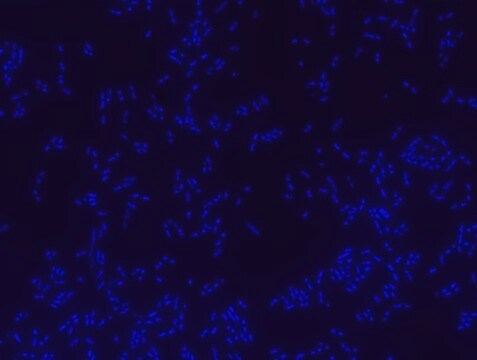
The yeast FISH (PF2-Cy3) probe specifically binds to a wide range of yeast species. Examples include Candida species that are common causes of bloodstream infections in humans, common environmental yeasts, and model organisms (Saccharomyces) in the phylum Ascomycota. The probe offered here was tested on 5 strains belonging to: Candida albicans, Candida boidinii, Candida glabrata, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. As a negative control, each slide contained fixed Proteus mirabilis bacterial cells in separate wells, to which the yeast FISH probe did not bind, confirming its specificity. For each experiment, an additional slide was stained with Eubacteria FISH probe - Cy3 (MBD0033). The Eubacteria FISH probe only stained the bacterial Proteus cells.
The yeast FISH (PF2) probe sequence was shown to specifically identify the yeast microbiome from various samples: murine intestine, blood samples and in environmental samples.
FISH can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (e.g., blood and other tissues), microbial ecology (biofilms, aquatic systems) and plants. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control probes (MBD0034/35) that accompany the specific probe of interest.
The yeast FISH (PF2-Cy3) probe specifically binds to a wide range of yeast species. Examples include Candida species that are common causes of bloodstream infections in humans, common environmental yeasts, and model organisms (Saccharomyces) in the phylum Ascomycota. The probe offered here was tested on 5 strains belonging to: Candida albicans, Candida boidinii, Candida glabrata, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. As a negative control, each slide contained fixed Proteus mirabilis bacterial cells in separate wells, to which the yeast FISH probe did not bind, confirming its specificity. For each experiment, an additional slide was stained with Eubacteria FISH probe - Cy3 (MBD0033). The Eubacteria FISH probe only stained the bacterial Proteus cells.
The yeast FISH (PF2) probe sequence was shown to specifically identify the yeast microbiome from various samples: murine intestine, blood samples and in environmental samples.
Application
Yeast FISH probe-Cy3 is suitable to use as a probe for fluorescence in- situ hybridization (FISH) to specifically recognize yeast cells.
Features and Benefits
- Visualize, identify, and isolate yeast cells. Observe native yeast populations in a variety of samples.
- Specific, sensitive, and robust identification of yeasts in bacterial mixed population.
- FISH can complement PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on yeast morphology.
- Identifies yeasts in clinical samples.
- The ability to detect yeasts in their natural habitat is an essential tool for studying host-microbiome interaction.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol
Alexandra J Scupham
Applied and Environmental Microbiology, 72, :793-:780 (2006)
Dominance of yeast in activated sludge under acidic pH and high organic loading
S.Zheng
Biochemical Engineering Journal, 52, Dominance of yeast in activated sludge under acidic pH and high organic loading-Dominance of yeast in activated sludge under acidic pH and high organic loading (2010)
V A Kempf et al.
Journal of clinical microbiology, 38(2), 830-838 (2000-02-03)
Using fluorescent in situ hybridization (FISH) with rRNA-targeted fluorescently labelled oligonucleotide probes, pathogens were rapidly detected and identified in positive blood culture bottles without cultivation and biotyping. In this study, 115 blood cultures with a positive growth index as determined
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service