Colony PCR
Colony PCR is a simple and quick method for verifying correct assembly of a cloned DNA construct. Traditionally, verification of DNA insertion during cloning was time-consuming, following bacterial transformation with laborious DNA purification and subsequent restriction enzyme digest. Colony PCR is a Direct PCR method that bypasses both DNA isolation and restriction digest, providing a fast, easy, and inexpensive solution for screening cloned constructs. Colony PCR applications are also compatible with screening clones created by sequence- and restriction-independent methods.
REDExtract-N-Amp™ PCR ReadyMix™ and JumpStart™ REDTaq® PCR ReadyMix™ for Colony PCR Applications
Suitable for use in colony PCR applications, REDExtract-N-Amp™ PCR ReadyMix™ (Cat. No. R4775) and JumpStart™ REDTaq® PCR ReadyMix™ (Cat. No. P0982) both provide buffer, polymerase, dNTPs, and stabilizers as a single 2x solution. They are formulated to allow for direct PCR, followed by loading of the samples onto an agarose gel after PCR without the need for additional buffers or loading dyes. The inert dye migrates at the same rate as a 125 base pair fragment, providing visual confirmation of DNA migration.
Features and Benefits
- Ultimate Flexibility: DNA purification before amplification is not necessary. Suitable for use with common strains of E. coli, such as SIG10 5a, NovaBlue (K-12), BL21(DE3), and DH5α strains, as well as S. pombe yeast. Sample can also be re-amplified, such as in nested PCR.
- High Specificity & Yield: JumpStart™ Taq DNA polymerase, an antibody inactivated hot start enzyme, is designed to minimize non-specific amplification while increasing target yield & specificity. JumpStart™ Taq amplifies targets up to 7 kb in length.
- Convenience: Hot start features in JumpStart™ products allows reaction set up at room temperature, which are beneficial when working with multiple samples. REDExtract-N-Amp™ PCR ReadyMix™ and JumpStart™ REDTaq® PCR ReadyMix™ are stable at 4 °C or −20 °C.
- Inhibitor Tolerance: JumpStart™ REDTaq® PCR ReadyMix™ has been shown to tolerate LB concentrations up to 20% and LB Agar or Terrific Broth up to 10%.
- Reproducibility: Reduces risk of contamination from multiple pipet steps and provides consistent performance across multiple reactions.
- Gel-ready: No loading buffer or tracking dye needs to be added. The PCR product is loaded directly into an agarose gel after amplification. The red tracer migrates at approximately the same rate as a 125bp fragment (slightly faster than bromophenol blue).
Colony PCR Protocol
The colony PCR protocol is straightforward: simply add a small amount of a colony from a plate or dense bacterial liquid culture to the PCR master mix and proceed to thermocycling. Cell lysis occurs during the initial high-temperature incubation. The resulting PCR products can be analyzed with gel electrophoresis or other DNA detection methods. This time-saving method is perfect for screening just a few colonies or for high throughput screenings of many clones.
1. Prepare PCR master mix (scaled based on the number of samples to analyze):
2. Dispense PCR master mix (20 µL) into each PCR tube or plate.
3. Using a sterile micropipette tip or a sterile toothpick transfer cells from each colony to a PCR tube and briefly stir to resuspend them in the PCR master mix. The mix may look slightly cloudy. Note: Don’t pick too many cells. Overloading cells will interfere with the PCR.
4. Amplify target DNA with the following thermocycling conditions:
Note: The cell lysis step is longer than a typical initial denaturation to efficiently break the bacterial wall
Colony PCR Applications with Bacteria and Yeast
Colony PCR was carried out using representative bacteria and yeast strains. E. coli competent cells SIG10 5a (Cat. No. CMC0001), NovaBlue (Cat. No. 69825), BL21(DE3) (Cat. No. 69450), and DH5α (Thermo Fisher Scientific) were transformed with pET28a(+) containing an inserted sequence. SIG10 5a was also transformed with an empty pET28a(+) vector. The E. coli colonies were screened for inserts using REDExtract-N-Amp™ PCR ReadyMix™ and flanking primers that amplify 1 kbp of the insert. All strains of E. coli were compatible with colony PCR, producing the target amplicons directly from the crude cell inputs without a DNA extraction step (Figure 1).

Figure 1Agarose gel of E. coli colony PCR products. Colony PCR of a 1 kbp insert was performed using REDExtract-N-Amp™ PCR ReadyMix™ and analyzed on a 1% agarose gel. Products were detected from all transformed cells containing pET28a(+) with inserts (samples 3-10), while no product was amplified from cells transformed with pET28a(+) empty vector (samples 1-2).
Schizosaccharomyces pombe yeast was transformed with pESP-1. PCR was performed on resulting colonies and plasmid control. The yeast colonies were screened for inserts using JumpStart™ REDTaq® PCR ReadyMix™ and flanking primers that amplify 226 bp of the insert. Like E. coli, the target sequences were amplified from yeast samples without DNA purification (Figure 2).

Figure 2Agarose gel of S. pombe colony PCR products. Colony PCR of a 226 bp insert was performed using JumpStart™ REDTaq® PCR ReadyMix™ and analyzed on a 1% agarose gel. Products were detected from all transformed cells (Colony 1-3) and plasmid template (Control).
Effect of Growth Media on PCR Yield Using JumpStart™ REDTaq® PCR ReadyMix™ Reagents
To quantify the impact of growth media contamination on PCR, common E. coli media LB, LB Agar, and Terrific Broth (TB) were spiked into JumpStart™ REDTaq® PCR ReadyMix™ reagents. Amplification was performed with pBR322 as a template, appropriate primers, α [32P] dCTP, and varying levels (0 – 50%) of LB, LB Agar, and TB. PCR yield was determined after trichloroacetic acid precipitation on glass filters followed by scintillation counting. Yields were normalized to 0% added media. Reactions were performed in triplicate. All of the media additions impacted product yield, but PCR using JumpStart™ REDTaq® PCR ReadyMix™ was still robust with up to 20% of the reaction mixture composed of LB and up to 10% of the reaction mixture composed of TB or LB agar (Figure 3). These levels exceed what would contaminate a typical colony PCR and demonstrate the high inhibitor tolerance of JumpStart™ REDTaq® PCR ReadyMix™ reagents.
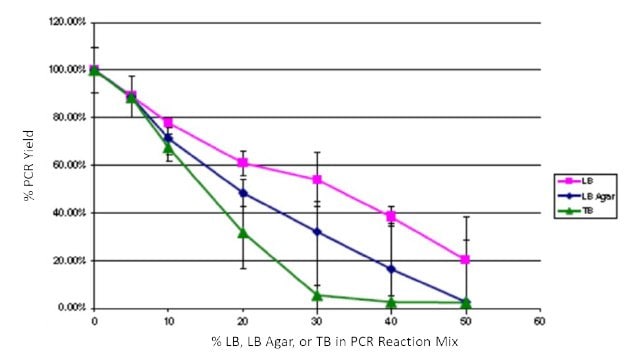
Figure 3Effects of spiking LB, LB Agar, and TB into PCR ReadyMix™ reagents. LB, pink squares; LB agar, blue diamonds; TB, green triangles.
Colony PCR Performance with JumpStartTM and REDTaq® ReadyMix™ Reagents
REDExtract-N-Amp™ PCR ReadyMix™, JumpStart™ REDTaq® PCR ReadyMix™, and other JumpStart™ and REDTaq® ReadyMix™ products, such as R2523 and P1107 are robust master mixes that allow for direct PCR of colonies and cultures from common cloning and expression strains, including E. coli and S. pombe, facilitating fast screening of cloned constructs. They have high tolerance to common PCR inhibitors, including LB concentrations up to 20% and LB Agar or Terrific Broth up to 10%, resulting in high performance in colony PCR. The master mixes can overcome the effects of cell debris and can be used directly with crude colonies or suspensions for the amplification of targets up to 7kb in length. Additional ultra-fast PCR master mixes, including our KOD One™ PCR Master Mix are also available and suitable for colony PCR applications. For additional PCR reagents and resources, explore our PCR Reagents and Kits resource hub.
Per continuare a leggere, autenticati o crea un account.
Non hai un Account?