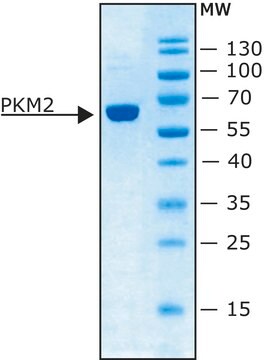
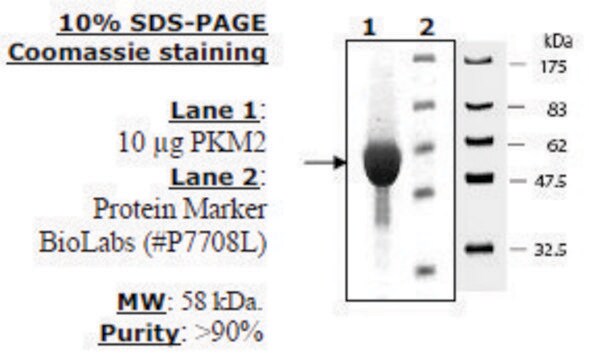
PK-RO
Roche
Pyruvate Kinase (PK)
from rabbit muscle
Synonym(s):
PK
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
Recommended Products
biological source
rabbit muscle
Quality Level
form
suspension
specific activity
~200 units/mg protein (at 25 °C (500 U/mg at 37 °C) with PEP as the substrate.)
packaging
pkg of 1 mL (10128155001 [10 mg])
pkg of 10 mL (10128163001 [100 mg])
manufacturer/tradename
Roche
optimum pH
7.0-7.5
storage temp.
2-8°C
Related Categories
General description
ATP:pyruvate 2-O-phosphotransferase
Pyruvate kinase has a molar mass of 237,000 and exists as a tetramer. Each polypeptide chain of this tetramer has a molar mass of 57,200. The enzyme contains two identical catalytic particles called protomers. Each of these protomers contains two polypeptide chains. Each protomer contains one site each for Mn2+ and phosphoenolpyruvate.
Pyruvate kinase has a molar mass of 237,000 and exists as a tetramer. Each polypeptide chain of this tetramer has a molar mass of 57,200. The enzyme contains two identical catalytic particles called protomers. Each of these protomers contains two polypeptide chains. Each protomer contains one site each for Mn2+ and phosphoenolpyruvate.
Application
Pyruvate kinase has been used to measure ATPase activity and in the determination of adenylate concentration.
Biochem/physiol Actions
Pyruvate kinase catalyzes the irreversible conversion of P-enolpyruvate and ADP to pyruvate and ATP with the utilization of a proton. The first step is the transfer of phosphate group from P-enolpyruvate to ADP with the formation of bound enolate of pyruvate and ATP. In the second step, a proton is added to enolate to generate the keto form of pyruvate. Apart from this, the enzyme exhibits other activities, such as ATP- and bicarbonate-dependent ATPase, phosphorylation of fluoride and hydroxylamine, ATP-dependent phosphorylation of glycolate, and decarboxylation of oxaloacetate.
Quality
Contaminants: <0.001% GK, <0.002% “NADH oxidase”, and ATPase, each, <0.01% enolase, LDH, and myokinase, each
Physical form
Suspension in 3.2 M ammonium sulfate solution, pH approximately 6
Preparation Note
Activator: PK requires Mg2+ (or Mn2+, Co2+) and K+ (or NH4+, Rb+) for full activity.
Other Notes
For life science research only. Not for use in diagnostic procedures.
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
No data available
Flash Point(C)
No data available
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Ilana M Nodelman et al.
EMBO reports, 14(12), 1098-1103 (2013-10-16)
Chromatin remodellers are ATP-dependent motor proteins that physically reposition and reorganize nucleosomes. Chd1 and Iswi-type remodellers possess a DNA-binding domain (DBD) needed for efficient nucleosome mobilization; however, it has not been clear how this domain physically contributes to remodelling. Here
Monika Ostaszewska et al.
Journal of plant physiology, 171(7), 549-558 (2014-03-25)
Sulphur, as a constituent of amino acids (cysteine and methionine), iron-sulphur clusters, proteins, membrane sulpholipids, glutathione, glucosinolates, coenzymes, and auxin precursors, is essential for plant growth and development. Absence or low sulphur concentration in the soil results in severe growth
T M Larsen et al.
Biochemistry, 33(20), 6301-6309 (1994-05-24)
The molecular structure of rabbit muscle pyruvate kinase, crystallized as a complex with Mn2+, K+, and pyruvate, has been solved to 2.9-A resolution. Crystals employed in the investigation belonged to the space group P1 and had unit cell dimensions a
Metabolic control and structure of glycolytic enzymes. 3. Dissociation and subunit structure of rabbit muscle pyruvate kinase.
M A Steinmetz et al.
Biochemistry, 5(4), 1399-1405 (1966-04-01)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service