LNA-based Probes
UPL is based on only 165 short hydrolysis probes. They are labeled at the 5' end with fluorescein (FAM) and at the 3' end with a dark quencher dye.
The extensive transcript coverage of the UPL probes is due to their short length of just 8 - 9 nucleotides and the selected sequences. In order to maintain the specificity and melting temperature (Tm) that hybridizing qPCR probes require, Locked Nucleic Acids (LNA) are incorporated into the sequence of each UPL probe. LNA's are DNA nucleotide analogues with increased binding strengths compared to standard DNA nucleotides.
The sequences of the 165 UPL probes have been carefully selected to detect 8- and 9-mer motifs that are very prevalent in the transcriptomes, ensuring optimal coverage of all transcripts in a given transcriptome.
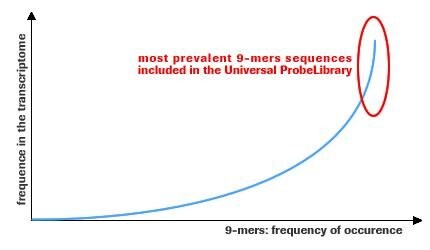
Figure 1.Frequency of 9-mer occurrence.
Within the human transcriptome, each probe binds to approximately 7,000 transcripts, while each transcript is detected by approximately 16 different probes. Only one specific transcript is detected at a time in a given PCR assay, as defined by the set of chosen PCR primers. For each assay, the design software suggests an optimal set of PCR primers, a probe, and any possible alternative sets.
UPL assays are compatible with all real-time PCR instruments capable of detecting fluorescein, FITC, FAM, and/or SYBR Green I. UPL assays have been used successfully on the LightCycler® 480 System, the LightCycler® Carousel-Based System, and other real-time PCR instruments from several suppliers.
LNA Technology
Locked Nucleic Acids (LNA) is a class of nucleic acids analogues, where the ribose ring is "locked" with a methylene bridge connecting the 2'-O atom with the 4'-C atom (Figure 2). LNA nucleosides containing the six common nucleobases (T, C, G, A, U and mC) that appear in DNA and RNA are able to form base-pairs with their complementary nucleosides according to the standard Watson-Crick base pairing rules. Therefore, LNA nucleotides can be mixed with DNA or RNA bases in the oligonucleotide whenever desired. The locked ribose conformation enhances base stacking and backbone pre-organization, this gives rise to an increased thermal stability and discriminative power of duplexes. LNA discriminates single base mismatches under conditions not possible with other nucleic acids (Figure 3).
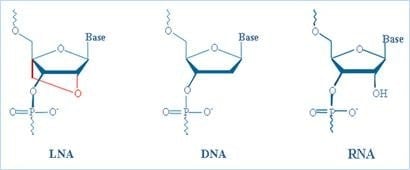
Figure 2.Comparison of LNA, DNA, and RNA.

Figure 3.Comparison of mismatch conditions for LNA and DNA.
Multiplex Assays
Multiplex Assays with Universal ProbeLibrary Reference Gene Assays
Use the Universal ProbeLibrary Reference Gene Assays together with the Universal ProbeLibrary to easily quantify expression levels of a human, mouse, or rat gene of interest in relation to an endogenous reference gene in a dual-color assay. Three reference gene assays are available for human (G6PDH, GAPD and ACTB- gene), 2 for mouse (ACTB-, and GAPD gene each).
Each Universal ProbeLibrary Reference Gene Assay provides a specifically designed 12-mer UPL reference gene probe and the corresponding reference gene-specific primer pair in a separate tube. The probe is labeled with LightCycler® Yellow 555 at the 5´ end and with a dark quencher dye near the 3´ end, to enable dual-color assays together with the standard UPL probes, which are labeled with FAM. The UPL reference gene probes can be detected using real-time PCR instruments with excitation filters of 470 nm to 530 nm and emission filters of 550 nm to 610 nm. In multiplex assays, a standard FAM or SYBR Green I filter is used as the first channel to detect FAM-labeled UPL probes for the gene of interest, and a longer-wavelength emission filter (usually the next possible emission filter moving to longer wavelengths, e.g.,560 nm or 568 nm) is used as the second channel to detect UPL reference gene probes.
Multiplex PCR assays for a target gene and a UPL Reference Gene are designed by the ProbeFinder software at the Assay Design Center with a high success rate of over 90%. In case no matching assays are found for the reference gene of choice (out of the listed assays), ProbeFinder SW suggests dual-color assays with one or more other reference genes out of the list.
Designing multiplex assays is as easy as designing monoplex assays: when you select the multiplex option online, ProbeFinder will design multiple UPL assays for your gene of interest. At the same time, each assay is subjected to in silico PCR testing to verify that it can be multiplexed with one (or more) of the UPL Reference Gene Assays for the relevant organism.
Using the LightCycler® 480 Real-Time PCR System, the expression of 45 genes is detected with a Universal ProbeLibrary dual color assay when testing only one primer and probe combination (examples are shown in Figure 4). The detected Cp values of dual-color assays differed by no more than 0 to 2.0 cycles from those detected with the single-color approach.
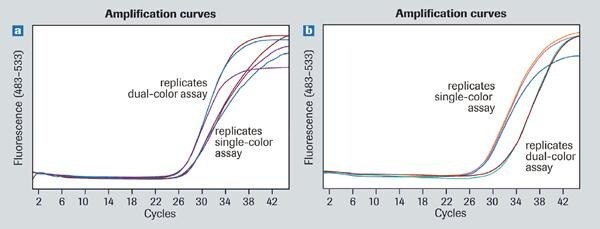
Figure 4.Expression of 45 genes detected with a Universal ProbeLibrary dual color assay.
Data kindly provided by Professor Stephan Wiemann, German Cancer Research Center, Heidelberg, Germany
如要继续阅读,请登录或创建帐户。
暂无帐户?