Ion Channel Flux Assays and Protocols
Ion Channel Assays for Measuring Ion Flux Through Potassium Channels
Ion channels are critical to regulating many signaling pathways in the cells, with multiple ion channels tuning electrical signals that are vital for a variety of cellular functions. Since ion channels play a key role in cellular signaling events, they are both drug targets and the sites of many off-target effects for drugs. Thus, assessment of how drugs modulate ion channel function is crucial for drug development and patient safety. This technical report provides a simple, high-throughput method for measuring ion flux through potassium ion channels, which is easily adaptable for sodium chloride and proton channels and has been utilized by many labs1-3. This assay, which may incorporate the use of artificial liposomes, can be employed in high-throughput screens to identify drug effects on ion channels1. It is also commonly used to assess functionality of purified membrane proteins used in structural studies and is the gold standard for determining lipid regulation of ion channels2. For additional information and ion channel product information, please visit our Ion Channels page or explore our Liposome Preparation resource page.
Fluorescence Sodium or Potassium Ion Channel Flux Assays
In this assay, purified ion channels are reconstituted into liposomes containing a high concentration (100-400 mM) of the conducting ion (K+, Na+). These liposomes are then diluted into a reaction buffer that external ion is osmotically compensated to generate an electrochemical gradient. A proton ionophore (CCCP) provides a path for protons to counter the efflux of ions in the presence of a conducting channel. Efflux of protons is then monitored by the membrane-permeable pH-sensitive dye ACMA. At neutral pH, ACMA has bright fluorescence. However, upon protonation, ACMA quenches and is no longer membrane-permeable.

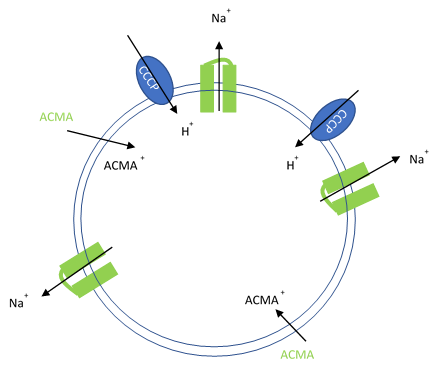
Figure 1. Artificial liposomes containing proton ionophores (CCCP) for protons to counter the efflux of potassium or sodium ions and membrane permeable sensitive dye ACMA is utilized to monitor the efflux of protons.
After ion and proton flux have reached an equilibrium (5-30 minutes), available liposome volume is measured by the addition of valinomycin or monensin, which provides a channel-independent path for K+ or Na+ conduction. Molecules or lipids can be added when forming the liposomes to assess their activatory or inhibitory affect. Example data can be found in Figure 2 with the potassium selective Kirbac and cation-selective NaK channel.


Figure 2. Sample data utilizing fluorescence-based flux assays to investigate either potassium or sodium flux in artificial liposomes.
The potassium or sodium flux sample data demonstrate the coupling of an internal change of pH to the flux of an ion through a pore, and are easily adjusted to test for ion conduction of many types depending on the channel reconstituted. For example, liposomes reconstituted with high Na can be used to test for Na channel activity. This assay can be used for K, Na, H, Cl channels by using the correct ionophores and adjusting the order the CCCP and ionophores are added.
Proteoliposome Preparation
Lipids play a key role in regulating ion channels, but this regulation is difficult to quantitatively assess in a cellular context. Proteoliposomes formed in this assay can be made with a variety of lipids allowing for quantitative experiments on lipid regulation of ion channels4. For additional resources, please visit our liposome preparation page.
Liposome Formation Methods
There are a number of ways in which liposomes may be formed depending on the following needs: Batch size, lipid conditions, storage requirements, and protein activity. The three most common ways to form liposomes are by dialysis, Amberlite® resin or gel filtration, with each of the three providing unique advantages and disadvantages.
Dialysis: Dialyzing using a small molecular weight cutoff allows the small free detergent molecules to exchange with the larger dialysis volume (1000X dialysis). Additionally, since lipids do not exchange in solution, they will remain in the dialysis tubing forming proteoliposomes. It is recommended that dialysis is performed above the lipids transition temperature. This method works best for detergents with high CMC and is less effective for detergents like Triton X-100 and DDM with very low CMC’s. While this is considered a gentle method of reconstitution and may work well with fragile proteins, it is a time-consuming process with several buffer changes required and generally requires an overnight step. Rate of detergent removal is hard to control and may make for less reproducible liposome.
Amberlite®: Polystyrene beads (Amberlite® XAD®-2) containing small hydrophobic pores absorb a wide variety of detergents and are commonly used for liposome preparation5. The amount of Amberlite® added determines the rate of detergent absorption and is generally added in small batches1, with approximately 30 mg beads/mg detergent total. Some lipids and proteins may be absorbed into the Amberlite®, however their large size prevents significant absorption. This method is relatively fast with absorption completed within hours. Addition of the same ratio of Amberlite® to detergent will result in a consistent liposome formation. This method is amendable to small and large batch sizes and relatively easy to generate samples in parallel.
Gel Filtration: This method is the fastest and most efficient way to form proteoliposomes, with samples ready within 20min of starting, making it useful when many samples are needed2. Large liposomes pass quickly through the Sephadex G-25 columns, whereas the free detergents are slowed by diffusing into the pores. Using commercially available Sephadex columns aids in generating consistent liposomes, as the size of the column plays an important role in the final proteoliposome size and purity. One downside is that the sample is diluted during the run, so if the exact protein concentration needs to be known, it will need to be quantified. Also, some lipids are absorbed into the column.
Liposome Formation using Gel Filtration and Fluorescence-Based Flux Assay Protocol Reagents
- PD SpinTrap™ G-25 or 96 well plate
- Lipid Solubilizing buffer: 35 mM Chaps, 450 mM KCl , 10 mM HEPES, 4 mM NMDG, and 0.5 mM EGTA, pH 7.4
- Flux inside buffer: 20 mM Hepes, 1 mM EDTA, 450 mM KCl, pH 7.4
- Flux outside buffer: 20 mM Hepes, 1mM EDTA, 450 mM NMDG
- 1000X ACMA stock: 2 mM ACMA in DMSO
- 200X 0.4 mM CCCP in DMSO
- 200X 4 uM Valinomycin in DMSO
- 200X 5 mM Monensin in DMSO
- Black 96 well plate
Liposome Formation Gel Filtration Protocol
- Equilibrate the PD SpinTrapTM G25 columns as directed with Flux inside buffer (20 mM Hepes, 1 mM EDTA, 450 mM KCl, pH 7.4)
- Mix 1-10 μg of 1 mg/mL solubilized membrane protein with 100 μL 10 mg/mL POPE:POPG solubilized in 35 mM Chaps ,450 mM KCl, 10 mM HEPES, 4 mM NMG, and 0.5 mM EGTA, pH 7.4 and let equilibrate for 20 min on ice or at room temperature. Also add the same volume of detergent containing buffer with the membrane protein to the 10 mg/mL POPE:POPG to create an empty liposome control.
- Dry the Sephadex® column by spinning to up to 800g 1 min.
- Load 100 μL of protein:detergent mixture onto the center of the equilibrated PD Spintrap G25 column. Add 40 μL stacker flux inside buffer.
- Spin loaded spin column 800xg for 2 min and collect the flow through which contains loaded proteoliposomes in high K buffer.
Fluorescence-based flux assay protocol
- Load 10 μL proteoliposomes containing high ion concentration onto 96-well plate
- Add 10 μL 1000X ACMA to 10mL of flux outside buffer
- Add 190 μL Flux outside buffer to the black 96-well plate containing proteoliposomes and mix.
- Collect 10 time points of initial ACMA fluorescence, excitation 419 nm emission 490 nm
- Add 1 μL of 200X CCCP and quickly mix and collect fluorescence until flux has equilibrated (5-30 min)
- Add 1 μL 200X Valinomycin (potassium) or Monensin (sodium) mix and quickly collect fluorescence for 10 time points
- Calculate total flux with this equation
Calculate total flux with this equation
Flux= (Flux at time point N – Flux Final) / (Flux Initial - Flux Final)
Optimization and Storage of Proteoliposomes:
Proteins embedded in a lipid bilayer are generally much more stable than in detergents, which allows for long-term storage of the proteoliposomes. Many proteins in proteoliposomes may be frozen and stored at -80 °C for long periods to time, but it is best to test for loss of function after thawing. The proteoliposomes formed by the three detergent removal methods are of mixed size and may be a mixture of multilamellar and unilamellar vesicles. The flux assay’s signal is dependent upon the size and unilamellar nature of the liposomes, with larger unilamellar vesicles having larger flux capacity. If the channels used in these experiments have low flux or conductance it is advised to do some post liposome formation processing that is outlined below.
Freeze Thaw:
To form unilamellar proteoliposomes liposomes are quickly frozen (generally in liquid nitrogen) and thawed above their transition temperature. This process can be repeated several times, with each freeze thaw cycle leading to smaller but more unilamellar proteoliposomes. Freezing and thawing work particularly well for experiments that require many samples as it can be done in parallel.
Extrusion:
Unilamellar proteoliposomes can be formed by forcing them through polycarbonate filters with a defined pore size. This can be done with the LiposoFast Liposome Factory with pores from 100-400 nm. However, this process is time consuming and likely not conducive to experiments with many individual liposomes.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?