All Photos(1)
About This Item
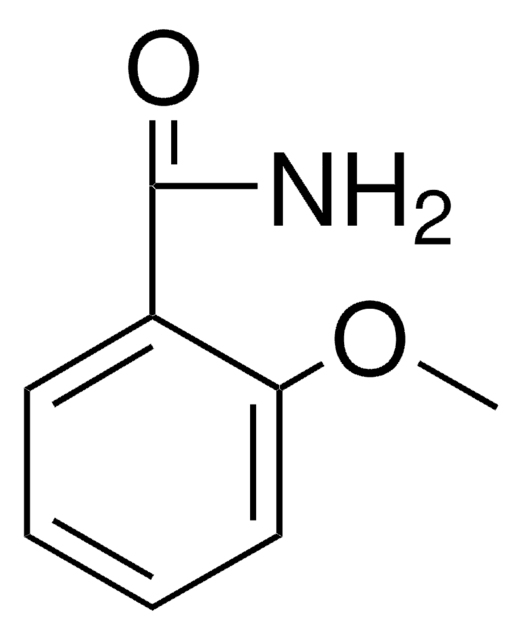
Linear Formula:
CH3OC6H4CONH2
CAS Number:
Molecular Weight:
151.16
Beilstein:
2206857
EC Number:
MDL number:
UNSPSC Code:
12352100
PubChem Substance ID:
NACRES:
NA.22
Recommended Products
Quality Level
Assay
97%
mp
132.5-135.5 °C (lit.)
SMILES string
COc1cccc(c1)C(N)=O
InChI
1S/C8H9NO2/c1-11-7-4-2-3-6(5-7)8(9)10/h2-5H,1H3,(H2,9,10)
InChI key
VKPLPDIMEREJJF-UHFFFAOYSA-N
Looking for similar products? Visit Product Comparison Guide
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
S Hauschildt et al.
Advances in experimental medicine and biology, 419, 249-252 (1997-01-01)
Stimulating monocytes/macrophages with bacterial lipopolysaccharide (LPS) results in TNF-alpha, IL-1, IL-6 and nitrite (NO2-) formation. Inhibitors of poly(ADP-ribose)polymerase inhibit release of these mediators by preventing mRNA expression indicating that ADP-ribosylation plays a crucial role in the synthesis of these mediators.
Y Ohashi et al.
Journal of bacteriology, 181(4), 1348-1351 (1999-02-11)
3-Methoxybenzamide (3-MBA), which is known to be an inhibitor of ADP-ribosyltransferase, inhibits cell division in Bacillus subtilis, leading to filamentation and eventually lysis of cells. Our genetic analysis of 3-MBA-resistant mutants indicated that the primary target of the drug is
G Golderer et al.
The Biochemical journal, 277 ( Pt 3), 607-610 (1991-08-01)
ADP-ribosylation of core histones was investigated in isolated nuclei of Physarum polycephalum. Core histone species differed in the mode of modification. Whereas ADP-ribosylation of H2A and H2B is sensitive to inhibition by 3-methoxybenzamide, as with most other nuclear acceptor proteins
N Nemoto et al.
Carcinogenesis, 12(4), 623-629 (1991-04-01)
Regulation of P(1)450 gene expression in mouse hepatocytes from responsive (C57BL/6) and non-responsive (DBA/2) strains in primary culture was investigated with respect to aryl hydrocarbon hydroxylase (AHH) activity and P450 transcript levels. Although significant induction of AHH activity in C57BL/6
H Altmann
Acta biologica Hungarica, 41(1-3), 19-34 (1990-01-01)
Poly(ADP-ribose)-polymerase is an important cellular regulatory enzyme which can change chromatin structure and function. Action mechanisms and activation of the enzyme are described. The synthesis of poly(ADP-ribose) can be modulated by interaction of substances with the DNA binding site of
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service