All Photos(1)
About This Item
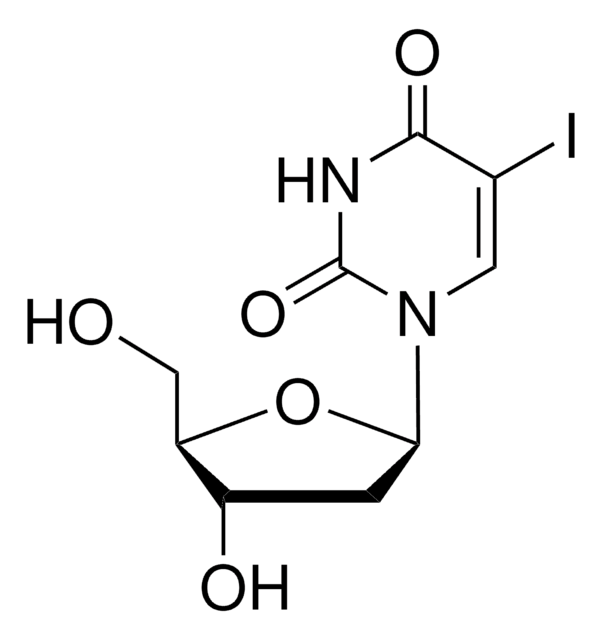
Empirical Formula (Hill Notation):
C11H12N2O6
CAS Number:
Molecular Weight:
268.22
UNSPSC Code:
41106305
NACRES:
NA.22
Recommended Products
Assay
≥95%
form
powder
reaction suitability
reaction type: click chemistry
storage temp.
−20°C
InChI
1S/C11H12N2O6/c1-2-5-3-13(11(18)12-9(5)17)10-8(16)7(15)6(4-14)19-10/h1,3,6-8,10,14-16H,4H2,(H,12,17,18)/t6-,7-,8-,10-/m1/s1
InChI key
QCWBIPKYTBFWHH-FDDDBJFASA-N
Related Categories
Application
This alkyne-bearing metabolic labeling reagent can be used to measure RNA synthesis in proliferating cells. Once fed to cells, this nucleoside is incorporated during active RNA synthesis and can then be detected using a copper-catalyzed click reaction with fluorescent dyes. The cells can then be analysed by fluorescence imaging, flow cytometry or high content imaging analysis.
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Cindy Y Jao et al.
Proceedings of the National Academy of Sciences of the United States of America, 105(41), 15779-15784 (2008-10-09)
We describe a chemical method to detect RNA synthesis in cells, based on the biosynthetic incorporation of the uridine analog 5-ethynyluridine (EU) into newly transcribed RNA, on average once every 35 uridine residues in total RNA. EU-labeled cellular RNA is
Christophe Bourges et al.
EMBO molecular medicine, 12(5), e12112-e12112 (2020-04-03)
Deriving mechanisms of immune-mediated disease from GWAS data remains a formidable challenge, with attempts to identify causal variants being frequently hampered by strong linkage disequilibrium. To determine whether causal variants could be identified from their functional effects, we adapted a
Xichen Bao et al.
Nature methods, 15(3), 213-220 (2018-02-13)
We combine the labeling of newly transcribed RNAs with 5-ethynyluridine with the characterization of bound proteins. This approach, named capture of the newly transcribed RNA interactome using click chemistry (RICK), systematically captures proteins bound to a wide range of RNAs
Magdalena P Crossley et al.
Nucleic acids research, 48(14), e84-e84 (2020-06-17)
R-loops are dynamic, co-transcriptional nucleic acid structures that facilitate physiological processes but can also cause DNA damage in certain contexts. Perturbations of transcription or R-loop resolution are expected to change their genomic distribution. Next-generation sequencing approaches to map RNA-DNA hybrids
Kaishun Hu et al.
Advanced science (Weinheim, Baden-Wurttemberg, Germany), 7(20), 2000157-2000157 (2020-10-27)
Repair of DNA double-strand breaks (DSBs) is essential for genome integrity, and is accompanied by transcriptional repression at the DSB regions. However, the mechanisms how DNA repair induces transcriptional inhibition remain elusive. Here, it is identified that BRD7 participates in
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service


![Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine 97%](/deepweb/assets/sigmaaldrich/product/structures/179/695/86a721c8-2a4c-4e4f-bc36-6276ce7a941f/640/86a721c8-2a4c-4e4f-bc36-6276ce7a941f.png)