All Photos(1)
About This Item
Empirical Formula (Hill Notation):
C9H17N3O4
CAS Number:
Molecular Weight:
231.25
EC Number:
MDL number:
UNSPSC Code:
12352209
PubChem Substance ID:
NACRES:
NA.26
Recommended Products
Product Name
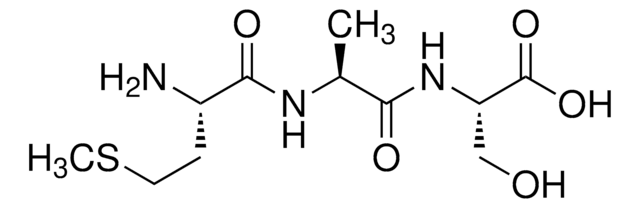
Ala-Ala-Ala,
Assay
≥98% (TLC)
Quality Level
form
powder
color
white
storage temp.
−20°C
SMILES string
C[C@H](N)C(=O)N[C@@H](C)C(=O)N[C@@H](C)C(O)=O
InChI
1S/C9H17N3O4/c1-4(10)7(13)11-5(2)8(14)12-6(3)9(15)16/h4-6H,10H2,1-3H3,(H,11,13)(H,12,14)(H,15,16)/t4-,5-,6-/m0/s1
InChI key
BYXHQQCXAJARLQ-ZLUOBGJFSA-N
Looking for similar products? Visit Product Comparison Guide
Amino Acid Sequence
Ala-Ala-Ala
Application
Trialanine (Ala-Ala-Ala) may be used along with other short chain alanines, tetra- and penta-alanine, as model compounds to study physicochemical parameters of small peptides.
Storage Class Code
11 - Combustible Solids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Kaicong Cai et al.
Physical chemistry chemical physics : PCCP, 11(40), 9149-9159 (2009-10-09)
A molecular mechanics (MM) force field-based empirical electrostatic potential map (MM map) for amide-I vibrations is developed with the aim of seeking a quick and reasonable approach to computing local mode parameters and their distributions in solution phase. Using N-methylacetamide
Reinhard Schweitzer-Stenner
The journal of physical chemistry. B, 113(9), 2922-2932 (2009-02-27)
The conformational preference of individual amino acid residues in the unfolded state of peptides and proteins is the subject of a continuous debate. Research has mostly been focused on alanine, owing to its abundance in proteins and its relevance for
Wenfei Li et al.
The Journal of chemical physics, 130(21), 214108-214108 (2009-06-11)
Biomolecular systems are inherently hierarchic and many simulation methods that try to integrate atomistic and coarse-grained (CG) models have been proposed, which are called multiscale simulations. Here, we propose a new multiscale molecular dynamics simulation method which can achieve high
Mandy C Green et al.
The Journal of chemical physics, 138(7), 074111-074111 (2013-03-01)
An open-shell extension of the pair interaction energy decomposition analysis (PIEDA) within the framework of the fragment molecular orbital (FMO) method is developed. The open-shell PIEDA method allows the analysis of inter- and intramolecular interactions in terms of electrostatic, exchange-repulsion
Jos Oomens et al.
Journal of the American Society for Mass Spectrometry, 20(2), 334-339 (2008-11-18)
Infrared multiple photon dissociation (IRMPD) spectroscopy is used to identify the structure of the b(2)(+) ion generated from protonated tri-alanine by collision induced dissociation (CID). The IRMPD spectrum of b(2)(+) differs markedly from that of protonated cyclo-alanine-alanine, demonstrating that the
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service