About This Item
Código UNSPSC:
41106609
Productos recomendados
aplicaciones
CRISPR
Condiciones de envío
dry ice
temp. de almacenamiento
−20°C
Categorías relacionadas
Descripción general
All-in-one, ready-to-use Cas9 and guide RNA (gRNA) expression plasmids for use with monocots and dicots.
CRISPR Plant Cas9 products are intended for Agrobacterium-mediated plant transformation or biolistic microparticle bombardment or protoplast transformation. The products are based on the type IIA CRISPR-Cas9 derived from Streptococcus pyogenes. The native Cas9 coding sequence is codon optimized for expression in monocots and dicots, respectively. The monocot Cas9 constructs contain a monocot U6 promoter for sgRNA expression, and the dicot Cas9 constructs contain a dicot U6 promoter.
The plant selection markers include:
CRISPR Plant Cas9 products are intended for Agrobacterium-mediated plant transformation or biolistic microparticle bombardment or protoplast transformation. The products are based on the type IIA CRISPR-Cas9 derived from Streptococcus pyogenes. The native Cas9 coding sequence is codon optimized for expression in monocots and dicots, respectively. The monocot Cas9 constructs contain a monocot U6 promoter for sgRNA expression, and the dicot Cas9 constructs contain a dicot U6 promoter.
The plant selection markers include:
- hygromycin B resistance gene
- neomycin phosphotransferase gene
- bar gene (phosphinothricin acetyl transferase)
Aplicación
- Inactivation of genes
- Target validation
- Site specific integration of gene of interest
- Gene replacement via HR
Características y beneficios
- Main advantages of CRISPR/Cas9 are in terms of simplicity, accessibility, cost and versatility.
- CRISPR/Cas9 system does not require any protein engineering steps, making it much more straightforward to test multiple gRNAs for each target gene
- Only 20 nt in the gRNA sequence need to be changed to confer a different target specificity
- Another advantage of CRISPR/Cas9 compared to ZFNs and TALENs is the ease of multiplexing. The simultaneous introduction of DSBs at multiple sites can be used to edit several genes at the same time. It can be particularly useful to knock out redundant genes or parallel pathways. Multiplex editing with the CRISPR/Cas9 system simply requires the monomeric Cas9 protein and any number of different sequence-specific gRNAs.
- CRISPR/Cas9 system can cleave methylated DNA in human cells allowing genomic modifications that are beyond the reach of the other nucleases. This has not been specifically explored in plants, it is reasonable to assume that the ability to cleave methylated DNA is intrinsic to the CRISPR/Cas9 system and not dependent on the target genome
Componentes
1 vial containing 50ul of 20ng/ul plasmid DNA
Keep reagent tubes closed when not in use.
Practice aseptic lab technique to avoid DNase contamination.
Keep reagent tubes closed when not in use.
Practice aseptic lab technique to avoid DNase contamination.
Principio
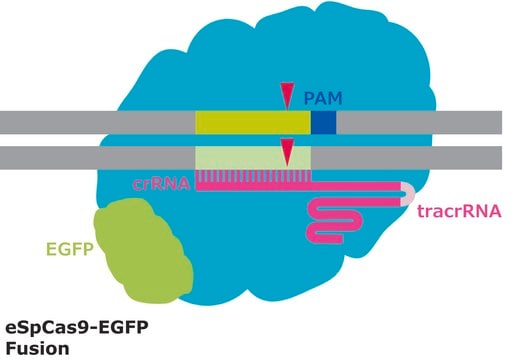
CRISPR/Cas systems are employed by bacteria and archaea as a defense against invading viruses and plasmids. Recently, the type II CRISPR/Cas system from the bacterium Streptococcus pyogenes has been engineered to function in eukaryotic systems using two molecular components: a single Cas9 protein and a non-coding guide RNA (gRNA). The Cas9 endonuclease can be programmed with a single gRNA, directing a DNA double-strand break (DSB) at a desired genomic location. Similar to DSBs induced by zinc finger nucleases (ZFNs), the cell then activates endogenous DNA repair processes, either non-homologous end joining (NHEJ) or homology-directed repair (HDR), to heal the targeted DSB.
Otras notas
For ordering any of our custom CRISPR plant products please visit: CUSTOM ORDERING FORM
Información legal
Producto relacionado
Código de clase de almacenamiento
12 - Non Combustible Liquids
Clase de riesgo para el agua (WGK)
WGK 1
Punto de inflamabilidad (°F)
Not applicable
Punto de inflamabilidad (°C)
Not applicable
Certificados de análisis (COA)
Busque Certificados de análisis (COA) introduciendo el número de lote del producto. Los números de lote se encuentran en la etiqueta del producto después de las palabras «Lot» o «Batch»
¿Ya tiene este producto?
Encuentre la documentación para los productos que ha comprado recientemente en la Biblioteca de documentos.
Los clientes también vieron
Nuestro equipo de científicos tiene experiencia en todas las áreas de investigación: Ciencias de la vida, Ciencia de los materiales, Síntesis química, Cromatografía, Analítica y muchas otras.
Póngase en contacto con el Servicio técnico