07-595
Anti-acetyl-Histone H4 (Lys12) Antibody
serum, Upstate®
Synonym(s):
H4K12Ac, Histone H4 (acetyl K12)
About This Item
Recommended Products
biological source
rabbit
Quality Level
antibody form
serum
antibody product type
primary antibodies
clone
polyclonal
species reactivity
Saccharomyces cerevisiae, human
manufacturer/tradename
Upstate®
technique(s)
ChIP: suitable (ChIP-seq)
dot blot: suitable
western blot: suitable
isotype
IgG
NCBI accession no.
shipped in
dry ice
target post-translational modification
acetylation (Lys12)
Gene Information
human ... HIST2H4B(554313)
General description
Acetylation of histone H4 occurs at several different lysine positions in the histone tail and is performed by a family of enzymes known as Histone Acetyl Transferases (HATs).
Specificity
Immunogen
Application
Epigenetics & Nuclear Function
Histones
Quality
Target description
Physical form
Storage and Stability
Analysis Note
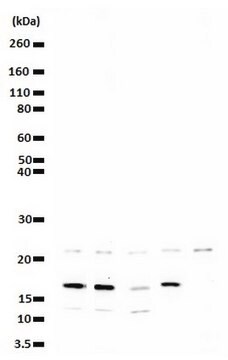
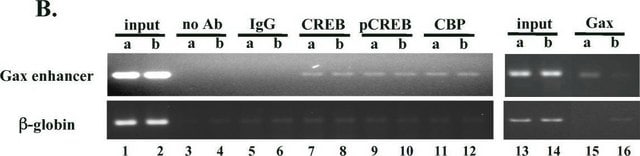
Acid extracted proteins from HeLa cells treated with sodium butyrate
Other Notes
Legal Information
Disclaimer
Not finding the right product?
Try our Product Selector Tool.
Storage Class Code
10 - Combustible liquids
WGK
WGK 1
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service