929441
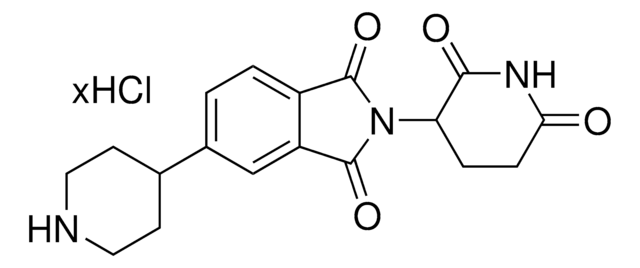
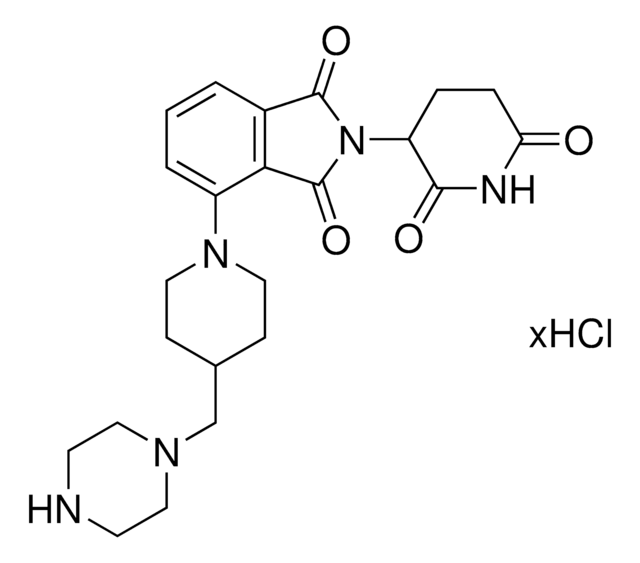
C5-Pomalidomide-piperazine hydrochloride
≥95%
Synonym(s):
2-(2,6-Dioxopiperidin-3-yl)-5-(piperazin-1-yl)isoindoline-1,3-dione hydrochloride
About This Item
Recommended Products
ligand
pomalidomide
Quality Level
Assay
≥95%
form
powder
storage temp.
2-8°C
SMILES string
O=C1N(C2CCC(NC2=O)=O)C(C3=CC=C(N4CCNCC4)C=C31)=O
Application
Technology Spotlight: Degrader Building Blocks for Targeted Protein Degradation
Protein Degrader Building Block
Other Notes
Legal Information
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Repr. 1B
Storage Class Code
6.1C - Combustible acute toxic Cat.3 / toxic compounds or compounds which causing chronic effects
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service