N8028
Nikkomycin Z from Streptomyces tendae
≥90% (HPLC)
Synonym(s):
Neopolyoxin C, Nikkomycin Z
About This Item
Recommended Products
Quality Level
Assay
≥90% (HPLC)
form
powder
solubility
H2O: soluble 5 mg/mL
antibiotic activity spectrum
fungi
Mode of action
cell wall synthesis | interferes
enzyme | inhibits
storage temp.
2-8°C
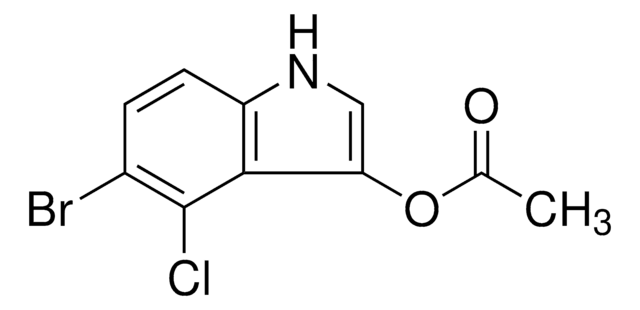
SMILES string
C[C@@H]([C@H](N)C(=O)N[C@H]([C@H]1O[C@H]([C@H](O)[C@@H]1O)N2C=CC(=O)NC2=O)C(O)=O)[C@H](O)c3ccc(O)cn3
InChI
1S/C20H25N5O10/c1-7(13(28)9-3-2-8(26)6-22-9)11(21)17(31)24-12(19(32)33)16-14(29)15(30)18(35-16)25-5-4-10(27)23-20(25)34/h2-7,11-16,18,26,28-30H,21H2,1H3,(H,24,31)(H,32,33)(H,23,27,34)/t7-,11-,12+,13-,14-,15+,16+,18+/m0/s1
InChI key
WWJFFVUVFNBJTN-JKEIIPFCSA-N
General description
Application
Biochem/physiol Actions
Other Notes
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Personal Protective Equipment
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service