MBD0042
Escherichia coli FISH probe - Cy3
Probe for fluorescence in situ hybridization (FISH)
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
UNSPSC Code:
12352200
NACRES:
NA.55
Recommended Products
Quality Level
technique(s)
FISH: suitable
fluorescence
λex 550 nm; λem 570 nm
shipped in
dry ice
storage temp.
−20°C
General description
Fluorescent In Situ Hybridization technique (FISH) is based on the hybridization of fluorescent labeled oligonucleotide probe to a specific complementary DNA or RNA sequence in whole and intact cells.1 Microbial FISH allows the visualization, identification and isolation of bacteria due to recognition of ribosomal RNA also in unculturable samples.2
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems 6) and plants7. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest.
Escherichia coli is a gram negative, facultative aerobic, rod-shaped coliform bacterium. E. coli colonizes the infant gut within hours of birth and establishes itself as the most abundant facultative anaerobe of the human intestinal microflora for the remainder of life, equipped with the abilities to grow in the ever-changing environment in the gut and cope with the mammalian host interaction.8,9 Nevertheless, E. coli can survive in many different ecological habitats, including abiotic environments, and is considered a highly versatile species. Known habitats of E. coli include soil, water, sediment, and food. Some strains of E. coli have evolved and adapted to a pathogenic lifestyle and can cause different disease pathologies.10
Escherichia coli probe specifically recognizes Escherichia coli cells.
Yet there are some reports that describe recognition of other bacteria with this probe, such as, Shigella boydii, Citrobacter davisae, Citrobacter lapagei, Citrobacter neteri 11 and Klebsiella pneumoniae 12.
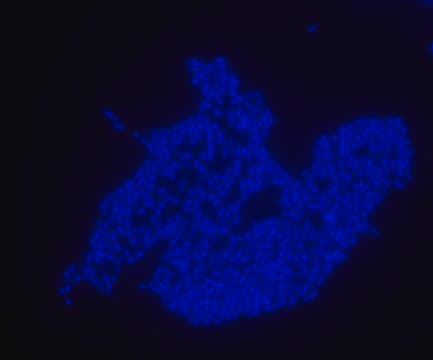
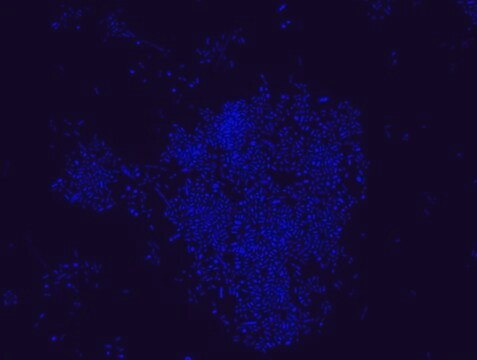
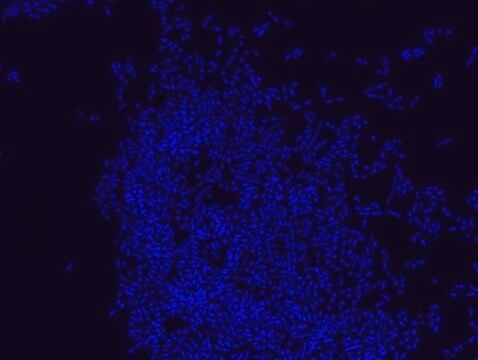
FISH technique was successfully used to identify E.coli with the probe in various samples such as pure culture (as described in the figure legends and11,13), large and small intestines samples14-16, fecal samples17-21, colonic biopsies18, urine samples, bladder and kidney sections embedded in paraffin22 and in E.coli biofilm23.
FISH technique can serve as a powerful tool in the microbiome research field by allowing the observation of native microbial populations in diverse microbiome environments, such as samples from human origin (blood3 and tissue4), microbial ecology (solid biofilms5 and aquatic systems 6) and plants7. It is strongly recommended to include positive and negative controls in FISH assays to ensure specific binding of the probe of interest and appropriate protocol conditions. We offer positive (MBD0032/33) and negative control (MBD0034/35) probes, that accompany the specific probe of interest.
Escherichia coli is a gram negative, facultative aerobic, rod-shaped coliform bacterium. E. coli colonizes the infant gut within hours of birth and establishes itself as the most abundant facultative anaerobe of the human intestinal microflora for the remainder of life, equipped with the abilities to grow in the ever-changing environment in the gut and cope with the mammalian host interaction.8,9 Nevertheless, E. coli can survive in many different ecological habitats, including abiotic environments, and is considered a highly versatile species. Known habitats of E. coli include soil, water, sediment, and food. Some strains of E. coli have evolved and adapted to a pathogenic lifestyle and can cause different disease pathologies.10
Escherichia coli probe specifically recognizes Escherichia coli cells.
Yet there are some reports that describe recognition of other bacteria with this probe, such as, Shigella boydii, Citrobacter davisae, Citrobacter lapagei, Citrobacter neteri 11 and Klebsiella pneumoniae 12.
FISH technique was successfully used to identify E.coli with the probe in various samples such as pure culture (as described in the figure legends and11,13), large and small intestines samples14-16, fecal samples17-21, colonic biopsies18, urine samples, bladder and kidney sections embedded in paraffin22 and in E.coli biofilm23.
Application
Probe for fluorescence in situ hybridization (FISH),recognizes Escherichia coli cells
Features and Benefits
- Visualize, identify and isolate Escherichia coli cells.
- Observe native E. coli cell populations in diverse microbiome environments.
- Specific, sensitive and robust identification of E. coli in bacterial mixed population.
- Specific, sensitive and robust identification even when E. coli is in low abundance in the sample.
- FISH can complete PCR based detection methods by avoiding contaminant bacteria detection.
- Provides information on E.coli morphology and allows to study biofilm architecture.
- Identify E.coli in clinical samples such as, urine samples, bladder and kidney sections (formalin-fixed paraffin-embedded (FFPE) samples), fecal samples and colon tissue.
- The ability to detect E.coli in its natural habitat is an essential tool for studying host-microbiome interaction.
Storage Class Code
12 - Non Combustible Liquids
WGK
nwg
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
S Favre-Bonté et al.
Infection and immunity, 67(11), 6152-6156 (1999-10-26)
The role of the Klebsiella pneumoniae capsular polysaccharide (K antigen) during colonization of the mouse large intestine was assessed with wild-type K. pneumoniae LM21 and its isogenic capsule-defective mutant. When bacterial strains were fed alone to mice, the capsulated bacteria
Eugenia Bezirtzoglou et al.
Anaerobe, 17(6), 478-482 (2011-04-19)
The development of the gut is controlled and modulated by different interacting mechanisms such as, genetic endowment, intrinsic biological regulatory functions, environment influences and last but no least, the diet influence. Considered together with other endogenous and exogenous factors the
Laure C Roger et al.
Microbiology (Reading, England), 156(Pt 11), 3317-3328 (2010-09-11)
From birth onwards, the gastrointestinal (GI) tract of infants progressively acquires a complex range of micro-organisms. It is thought that by 2 years of age the GI microbial population has stabilized. Within the developmental period of the infant GI microbiota
P V Kirjavainen et al.
Gut, 51(1), 51-55 (2002-06-22)
Recent data have outlined a relationship between the composition of the intestinal microflora and allergic inflammation, and demonstrated the competence of probiotics in downregulation of such inflammation. Our aims were to characterise the relationship between gut microbes and the extent
Sahar Soleimani et al.
Applied microbiology and biotechnology, 97(3), 1093-1102 (2012-09-11)
Biofilms of selected bacteria strains were previously used on metal coupons as a protective layer against microbiologically influenced corrosion of metals. Unlike metal surfaces, concrete surfaces present a hostile environment for growing a protective biofilm. The main objective of this
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service