A1757
6-Azauracil
≥98%
Synonym(s):
6-AU, 1,2,4-Triazine-3,5(2H,4H)-dione, 3,5-Dihydroxy-1,2,4-triazine, 6-Aza-2,4-dihydroxypyrimidine
Sign Into View Organizational & Contract Pricing
All Photos(2)
About This Item
Empirical Formula (Hill Notation):
C3H3N3O2
CAS Number:
Molecular Weight:
113.07
Beilstein:
116472
EC Number:
MDL number:
UNSPSC Code:
41106305
PubChem Substance ID:
NACRES:
NA.51
Recommended Products
Quality Level
Assay
≥98%
form
powder
mp
274-275 °C (lit.)
solubility
1 M NH4OH: 50 mg/mL, clear to slightly hazy, colorless to light yellow-green
SMILES string
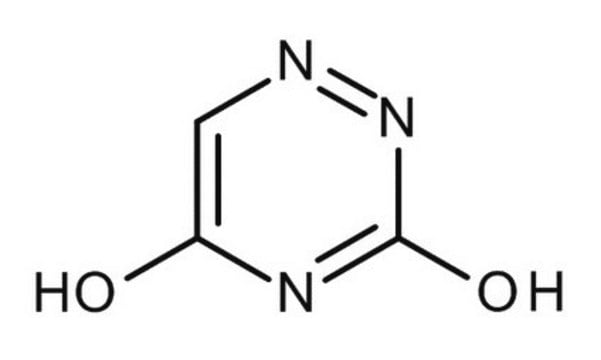
O=C1NN=CC(=O)N1
InChI
1S/C3H3N3O2/c7-2-1-4-6-3(8)5-2/h1H,(H2,5,6,7,8)
InChI key
SSPYSWLZOPCOLO-UHFFFAOYSA-N
Looking for similar products? Visit Product Comparison Guide
Application
6-Azauracil has been used as a transcriptional inhibitor to study its effects on the deletion of termination and polyadenylation protein (Tpa1) and Mag1 on cell viability. It has also been used as an orotidine-5′-monophosphate decarboxylase (OMPdecase) inhibitor in minimal media for determining the OMPdecase activity.
Biochem/physiol Actions
6-Azauracil (6-AU) is a pyrimidine analog of uracil and exhibits antitumor activity. It inhibits the growth of various microorganisms by depleting intracellular guanosine triphosphate (GTP) and uridine triphosphate (UTP) nucleotide pools.
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Peter L Freddolino et al.
eLife, 7 (2018-04-06)
Cells adapt to familiar changes in their environment by activating predefined regulatory programs that establish adaptive gene expression states. These hard-wired pathways, however, may be inadequate for adaptation to environments never encountered before. Here, we reveal evidence for an alternative
Kazuko Matsubara et al.
Genes to cells : devoted to molecular & cellular mechanisms, 12(1), 13-33 (2007-01-11)
The core histones are essential components of the nucleosome that act as global negative regulators of DNA-mediated reactions including transcription, DNA replication and DNA repair. Modified residues in the N-terminal tails are well characterized in transcription, but not in DNA
Clinical studies of 6-azauracil
Shnider BI, et al.
Cancer Research, 20(1), 28-33 (1960)
Hai-Ning Du et al.
Genes & development, 22(20), 2786-2798 (2008-10-17)
Set2-mediated H3 K36 methylation is an important histone modification on chromatin during transcription elongation. Although Set2 associates with the phosphorylated C-terminal domain (CTD) of RNA polymerase II (RNAPII), the mechanism of Set2 binding to chromatin and subsequent exertion of its
Kostyantyn V Dmytruk et al.
Metabolic engineering, 13(1), 82-88 (2010-11-03)
Currently, the mutant of the flavinogenic yeast Candida famata dep8 isolated by classic mutagenesis and selection is used for industrial riboflavin production. Here we report on construction of a riboflavin overproducing strain of C. famata using a combination of random
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service