MABE1095
Anti-DNA-RNA Hybrid Antibody, clone S9.6
clone S9.6, from mouse
Synonym(s):
DNA-RNA Duplex, DNA/RNA Duplex, DNA:RNA Duplex, DNA-RNA Hybrid, DNA/RNA Hybrid, DNA:RNA Hybrid, RNA-DNA Duplex, RNA/DNA Duplex, RNA:DNA Duplex, RNA-DNA Hybrid, RNA/DNA Hybrid, RNA:DNA Hybrid
About This Item
Recommended Products
biological source
mouse
Quality Level
antibody form
purified immunoglobulin
antibody product type
primary antibodies
clone
S9.6, monoclonal
species reactivity (predicted by homology)
all
technique(s)
ChIP: suitable
affinity binding assay: suitable
dot blot: suitable
immunocytochemistry: suitable
immunoprecipitation (IP): suitable
isotype
IgG2aκ
shipped in
ambient
target post-translational modification
unmodified
General description
Specificity
Immunogen
Application
Affinity Binding Assay: Clone S9.6 bound the DNA-RNA heteropolymer and poly(I)-poly(dC) equally, but 100-fold higher levels of poly(A)-poly(dT) were required to achieve a similar degree of binding. Single-stranded DNA, double-stranded DNA and RNA, and ribosomal RNA were not bound by clone S9.6 (Boguslawski, S.J., et al. (1986). J. Immunol Methods. 89(1):123-130).
Chromatin Immunoprecipitation (ChIP) Analysis: A representative lot detected increased DNA RNA hybrids at four actively transcribed genes upon shRNA-mediated knockdown of BRCA1 or BRCA2, but not PCID2 or RAD51 in HeLa cells (Bhatia, V., et al. (2014). Nature. 511(7509):362-365).
Chromatin Immunoprecipitation (ChIP) Analysis: A representative lot detected R-loops formed over beta-actin gene using HeLa chromatin preparation. RNase H treatment of the chromatin preparation prevented clone S9.6 from immunoprecipitating target chromatin fragments (Skourti-Stathaki, K., et al. (2011). Mol. Cell. 42(6):794-805).
Chromatin Immunoprecipitation-sequencing (ChIP-seq) Analysis: A representative lot detected genome-wide distribution of DNA-RNA hybrids in budding yeast by ChIP-seq analysis (El Hage, A., et al. (2014). PLoS Genet. 10(10):e1004716).
Immunocytochemistry Analysis: Representative lots immunolocalized nuclear R loops by fluorescent immunocytochemistry staining of methanol-fixed H1 human embryonic stem cells (hESCs) and formaldehyde-fixed HeLa cells (Bhatia, V., et al. (2014). Nature. 511(7509):362-365; Ginno, P.A., et al. (2012). Mol. Cell. 45(6):814-825).
Immunoprecipitation Analysis: A representative lot immunoprecipitated in vitro transcribed R-loop substrate (DNA-RNA hybrid), but not doouble-stranded DNA (dsDNA) (Ginno, P.A., et al. (2012). Mol. Cell. 45(6):814-825).
Epigenetics & Nuclear Function
Quality
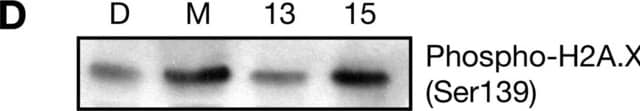
Immunocytochemistry Analysis: A 1:50 dilution of this antibody immunolocalized nuclear and mitochondrial DNA-RNA hybrids in 4% paraformaldehyde-fixed, 0.3% Triton X-100-permeabilized HeLa cells.
Physical form
Storage and Stability
Other Notes
Disclaimer
Not finding the right product?
Try our Product Selector Tool.
recommended
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 1
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Explore the basics of working with antibodies including technical information on structure, classes, and normal immunoglobulin ranges.
Antibodies combine with specific antigens to generate an exclusive antibody-antigen complex. Learn about the nature of this bond and its use as a molecular tag for research.
Learn differences in monoclonal vs polyclonal antibodies including how antibodies are generated, clone numbers, and antibody formats.
Immunofluorescence uses antibody-conjugated fluorescent molecules for protein localization, modification confirmation, and protein complex visualization.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service