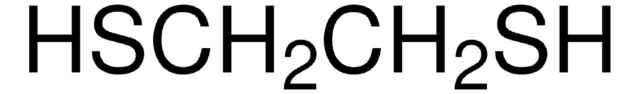77087
ω-Transaminase, Aspergillus fumigatus
recombinant, expressed in E. coli, ≥0.20 U/mg
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
CAS Number:
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.54
Recommended Products
recombinant
expressed in E. coli
Quality Level
form
powder
specific activity
≥0.20 U/mg
storage temp.
−20°C
Packaging
Bottomless glass bottle. Contents are inside inserted fused cone.
Unit Definition
1 U corresponds to the amount of enzyme which releases 1 μmol acetophenone per minute at 30°C. (R(−)-α-methyl-benzylamine as substrate).
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Resp. Sens. 1 - Skin Sens. 1
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
omega-Amino acid-pyruvate aminotransferase.
K Yonaha et al.
Methods in enzymology, 143, 500-504 (1987-01-01)
J S Shin et al.
Bioscience, biotechnology, and biochemistry, 65(8), 1782-1788 (2001-10-02)
Microorganisms that are capable of (S)-enantioselective transamination of chiral amines were isolated from soil samples by selective enrichment using (S)-alpha-methyl-benzylamine ((S)-alpha-MBA) as a sole nitrogen source. Among them, Klebsiella pneumoniae JS2F, Bacillus thuringiensis JS64, and Vibrio fluvialis JS17 showed good
Hyungdon Yun et al.
Applied and environmental microbiology, 70(4), 2529-2534 (2004-04-07)
Alcaligenes denitrificans Y2k-2 was obtained by selective enrichment followed by screening from soil samples, which showed omega-amino acid:pyruvate transaminase activity, to kinetically resolve aliphatic beta-amino acid, and the corresponding structural gene (aptA) was cloned. The gene was functionally expressed in
K Yonaha et al.
The Journal of biological chemistry, 267(18), 12506-12510 (1992-06-25)
The complete amino acid sequence of bacterial omega-amino acid:pyruvate aminotransferase (omega-APT) was determined from its primary structure. The enzyme protein was fragmented by CNBr cleavage, trypsin, and Staphylococcus aureus V8 digestions. The peptides were purified and sequenced by Edman degradation.
N Watanabe et al.
Journal of biochemistry, 105(1), 1-3 (1989-01-01)
The three-dimensional structure of omega-amino acid:pyruvate aminotransferase from Pseudomonas sp. F-126, an isologous alpha 4 tetramer containing pyridoxal 5'-phosphate (PLP) as a cofactor, has been determined at 2.0 A resolution. The diffraction data were collected with a newly developed Weissenberg
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service