C24604
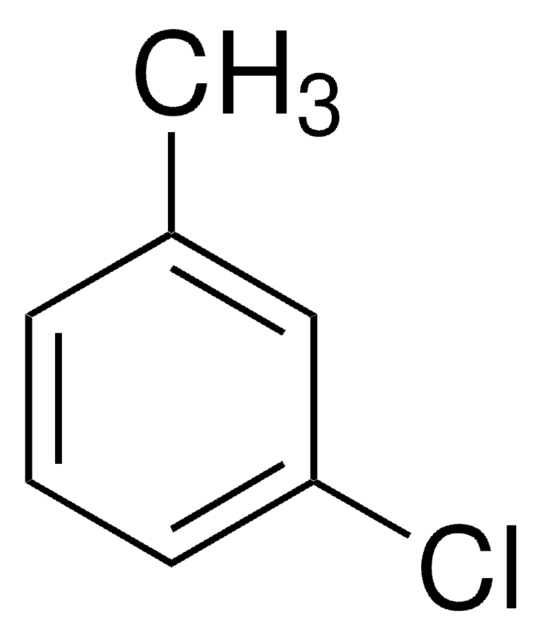
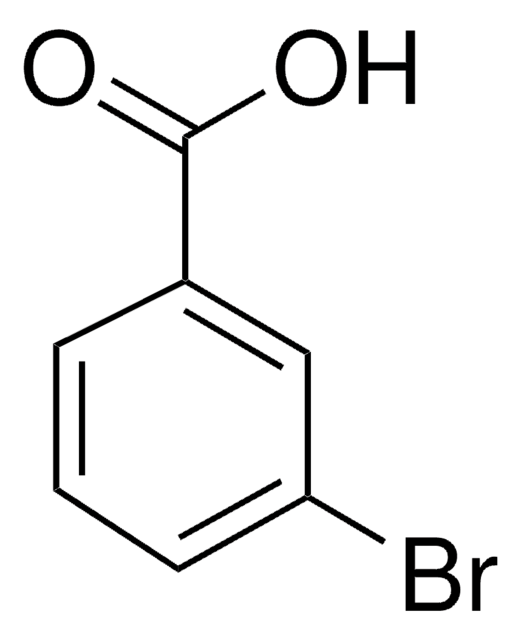
3-Chlorobenzoic acid
ReagentPlus®, ≥99%
Synonym(s):
m-Chlorobenzoic acid
Sign Into View Organizational & Contract Pricing
All Photos(3)
About This Item
Linear Formula:
ClC6H4CO2H
CAS Number:
Molecular Weight:
156.57
Beilstein:
907218
EC Number:
MDL number:
UNSPSC Code:
12352100
PubChem Substance ID:
NACRES:
NA.22
Recommended Products
product line
ReagentPlus®
Assay
≥99%
form
powder
mp
153-157 °C (lit.)
SMILES string
OC(=O)c1cccc(Cl)c1
InChI
1S/C7H5ClO2/c8-6-3-1-2-5(4-6)7(9)10/h1-4H,(H,9,10)
InChI key
LULAYUGMBFYYEX-UHFFFAOYSA-N
Looking for similar products? Visit Product Comparison Guide
Related Categories
Legal Information
ReagentPlus is a registered trademark of Merck KGaA, Darmstadt, Germany
Signal Word
Warning
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2 - Skin Irrit. 2
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Sudip K Samanta et al.
Molecular microbiology, 55(4), 1151-1159 (2005-02-03)
Rhodopseudomonas palustris strain RCB100 degrades 3-chlorobenzoate (3-CBA) anaerobically. We purified from this strain a coenzyme A ligase that is active with 3-CBA and determined its N-terminal amino acid sequence to be identical to that of a cyclohexanecarboxylate-CoA ligase encoded by
Alfredo Gallego et al.
World journal of microbiology & biotechnology, 28(3), 1245-1252 (2012-07-19)
An indigenous strain of Pseudomonas putida capable of degrading 3-chlorobenzoic acid as the sole carbon source was isolated from the Riachuelo, a polluted river in Buenos Aires. Aerobic biodegradation assays were performed using a 2-l microfermentor. Biodegradation was evaluated by
V P Jayachandran et al.
Journal of industrial microbiology & biotechnology, 36(2), 219-227 (2008-10-23)
The compatibility and efficiency of two ortho-cleavage pathway-following pseudomonads viz. the 3-chlorobenzoate (3-CBA)-degrader, Pseudomonas aeruginosa 3mT (3mT) and the phenol-degrader, P. stutzeri SPC-2 (SPC-2) in a mixed culture for the degradation of these substrates singly and simultaneously in mixtures was
S Yoshida et al.
Journal of applied microbiology, 106(3), 790-800 (2009-02-05)
To characterize biofilm formation of a chlorobenzoates (CBs) degrading bacterium, Burkholderia sp. NK8, with another bacterial species, and the biodegradation activity against CBs in the mixed-species biofilm. Burkholderia sp. NK8 was solely or co-cultured with each of five other representative
Juanita Larraín-Linton et al.
Journal of bacteriology, 188(19), 6793-6801 (2006-09-19)
Cupriavidus necator JMP134(pJP4) harbors a catabolic plasmid, pJP4, which confers the ability to grow on chloroaromatic compounds. Repeated growth on 3-chlorobenzoate (3-CB) results in selection of a recombinant strain, which degrades 3-CB better but no longer grows on 2,4-dichlorophenoxyacetate (2,4-D).
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service