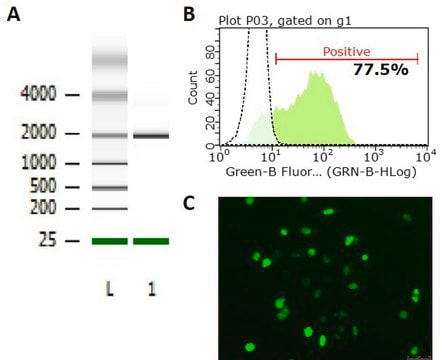
Due to the dimerization requirement of the FokI endonuclease, a pair of ZFNs is required to cause a double strand break. This strategy requires two different ZFNs to bind at the target site. Each ZFN recognizes a different 12-18 base pair target sequence, and these target sequences must be separated by 4-7 base pairs to allow formation of the catalytically active FokI dimer. These positional constraints drive a very high degree of specificity.
CSTZFN
CompoZr® Custom Zinc Finger Nuclease (ZFN)
ZFN plasmid and mRNA
Synonym(s):
Zinc Finger Nuclease
About This Item
Recommended Products
Looking for similar products? Visit Product Comparison Guide
Related Categories
General description
Application
- Creation of gene knockouts in multiple cell lines
- Complete knockout of genes not amenable to RNAi
- Creation of knock-in cell lines with promoters, fusion tags or reporters integrated into endogenous genes
- Creation of cell lines that produce higher yields of proteins or antibodies
Features and Benefits
- Rapid design, assembly, and validation of a ZFN pair targeting your gene of interest
- Rapid and permanent disruption of, or integration into, any genomic loci
- Mutations made are permanent and heritable
- Works in a variety of mammalian somatic cell types
- Edits induced through a single transfection experiment
- Knockout or knock-in cell lines in as little as two months
- Single or biallelic edits occur in 1-20% of clone population
- No antibiotic selection required for screening
Components
- Best Performing ZFN Pair
- 10 Aliquots of Ready-to-Deliver ZFN Pair in mRNA form
- ZFN Pair in Plasmid Form
- Forward and Reverse Primers that allow for determination of rate of mutation and for screening of individual clones
- Positive Control DNA
- Used to determine a baseline cutting efficiency
Other Notes
Custom ZFN Service offering HERE
Legal Information
related product
Storage Class Code
10 - Combustible liquids
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Choose from one of the most recent versions:
Certificates of Analysis (COA)
It looks like we've run into a problem, but you can still download Certificates of Analysis from our Documents section.
If you need assistance, please contact Customer Support.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
-
What specificity should be expected from a ZFN?
1 answer-
Helpful?
-
-
What is the Department of Transportation shipping information for this product?
1 answer-
Transportation information can be found in Section 14 of the product's (M)SDS.To access the shipping information for this material, use the link on the product detail page for the product.
Helpful?
-
-
What is a Zinc Finger Nuclease (ZFN)?
1 answer-
A ZFN is a hybrid molecule that couples the DNA binding domain of a zinc-finger protein with the DNA-cleaving nuclease domain of the restriction endonuclease FokI. The DNA binding motif specified by the zinc fingers directs the ZFN to a specific (targeted) locus in the genome.
Helpful?
-
-
Do the ZFNs remain in the cells after causing desired mutations?
1 answer-
No. The ZFNs are only expressed transiently but the genetic alteration is permanent. The transient nature of ZFNs also reduces the possibility of off-target effects.
Helpful?
-
-
How has the cleavage ability of my ZFN been tested?
1 answer-
To test the cleavage ability of the ZFNs we use a mismatch-specific nuclease assay. In this assay, PCR primers are used to amplify across the ZFN cleavage site in genomic DNA extracted from ZFN-treated cells. The resulting PCR products, which are now a mixture of wild type and mutated amplicons, are then denatured and re-annealed. When wild type and mutant alleles of the PCR products re-anneal with each other a mismatch occurs. The fragments are then exposed to a mismatch-specific nuclease and the digested reaction is run on a gel to look for smaller migrating cleavage products that indicate cleaved mismatches. The smaller migrating cleavage products reflect the frequency of mutated alleles and are a measure of in vivo ZFN efficacy. Quantitation of the released fragments allows a calculation of the percentage of mutated genes in the population.
Helpful?
-
-
I have a cell line I created by using a ZFN to knockout a gene. I would like to knockout a second gene in this cell line. Is this possible?
1 answer-
Yes, this is definitely a possibility. It has been shown that at least 3 different genes could be successively knocked out of the genome in the same cell line.
Helpful?
-
-
Can Zinc Finger Nucleases be synthesized to a specific mitochondrial DNA sequence?
1 answer-
This is target dependent, however the simple answer is yes- as long as sequence content is fairly diverse with some GC content. There is already literature for targeting of mtDNAs with ZFNs.
Helpful?
-
-
Should I expect off-target effects from my ZFN?
1 answer-
The ZFN reagents that we provide are unlikely to cause significant offtarget effects. The proprietary algorithm that Sigma uses to design the candidate ZFNs targets only sites that are unique within the genome. This, as well as the use of specially engineered obligate heterodimer FokI cleavage domains (Miller et al., Nature Biotechnology, July 25, 2007), helps guard against off-target effects for the ZFN you receive.
Helpful?
-
-
Will a ZFN insert a gene into my cell line?
1 answer-
Yes. To achieve targeted gene integration you must co-transfect your ZFN with a repair template, consisting of the gene to be inserted flanked by genomic sequence engineered to match (i.e., be homologous to) the sequence surrounding the intended ZFN cut site. After your ZFN cuts at the site specified by its sequence, the natural mechanism of homology dependent repair (HDR) takes over. The cell will recognize the homologous sequence in the repair template and insert the exogenous gene at the site of ZFN cleavage.
Helpful?
-
-
How is the binding specificity of the ZFN tested?
1 answer-
The binding specificity of the zinc finger modules in the Sangamo archive have previously been characterized by a number of in-vitro methods (ELISA among others).
Helpful?
-
Active Filters
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service